Hi all,
I am trying to fit a mutivariate time series model in numpyro. The model is a state-space model with AR(1). The data consists of two time series with missing values. I used the non-centered parametrization and jax.lax.scan as in Using lax.scan for time series in NumPyro. I am doing inference using NUTS. The generative process and the code of the model are the following:
![]()
![]()
![]()
![]()
![]()
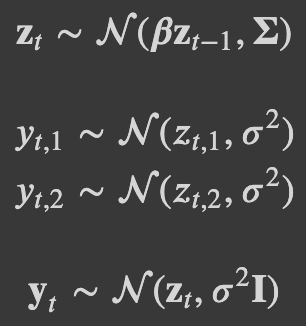
# Definition of the function for the jax.lax.scan
def f(carry, noise_t):
beta, z_prev = carry
z_t = beta*z_prev + noise_t
z_prev = z_t
return (beta, z_prev), z_t
def model(T, T_forecast, obs1=None, ix_mis1=None, ix_obs1=None, obs2=None, ix_mis2=None, ix_obs2=None):
"""
Prior for the betas, sigma and first state
"""
beta = numpyro.sample("beta", numpyro.distributions.Normal(jnp.zeros(2), jnp.ones(2)))
sigma = numpyro.sample("sigma", numpyro.distributions.HalfCauchy(jnp.ones(2)))
z_prev = numpyro.sample("z_prev", numpyro.distributions.Normal(jnp.zeros(2), jnp.ones(2)))
"""
Prior for the LKJ including a tau and the sampling noise
"""
tau = numpyro.sample("tau", numpyro.distributions.HalfCauchy(jnp.ones(2)))
L_Omega = numpyro.sample("L_Omega", numpyro.distributions.LKJCholesky(dimension=2, concentration=1.))
Sigma_lower = jnp.matmul(jnp.diag(jnp.sqrt(tau)), L_Omega)
noises = numpyro.sample("noises",
fn=dist.MultivariateNormal(loc=jnp.zeros(2), scale_tril=Sigma_lower),
sample_shape=(T+T_forecast-1,))
"""
Propagate the dynamics forward using jax.lax.scan
"""
carry = (beta, z_prev)
z_collection = [z_prev]
updated_carry, zs = lax.scan(f=f, init=carry, xs=noises, length=T+T_forecast-1)
z_collection = jnp.concatenate((jnp.array(z_collection), zs), axis=0)
"""
Sample the observed y (y_obs) and missing y (y_mis)
"""
y1_obs = numpyro.sample("y1_obs", numpyro.distributions.Normal(z_collection[ix_obs1, 0], sigma[0]), obs=obs1)
y1_mis = numpyro.sample("y1_mis", numpyro.distributions.Normal(z_collection[ix_mis1, 0], sigma[0]))
y1_pred = numpyro.sample("y1_pred", numpyro.distributions.Normal(z_collection[T:, 0], sigma[0]))
y2_obs = numpyro.sample("y2_obs", numpyro.distributions.Normal(z_collection[ix_obs2, 1], sigma[1]), obs=obs2)
y2_mis = numpyro.sample("y2_mis", numpyro.distributions.Normal(z_collection[ix_mis2, 1], sigma[1]))
y2_pred = numpyro.sample("y2_pred", numpyro.distributions.Normal(z_collection[T:, 1], sigma[1]))
I have two questions about the model:
- Why does the model “learn” a distribution for each of the noises? I would expect the posterior of noises to be just standard gaussians. Almost all of them are, but some are clearly not. If the noise is “exogenous” why can’t I input just numpy samples from a standard gaussian?
- When I run the model without the predictions (y1_pred and y2_pred) the model runs relatively smoothly (still with the problem in 1) but I get sensible results), but whenever I include the forecasting part the model collapses. That is, the posterior become single values sampled over and over with n_eff=0.5 for all of them.
Thank you,
Sergio