I’m seeking advice on improving runtime performance of the below numpyro model.
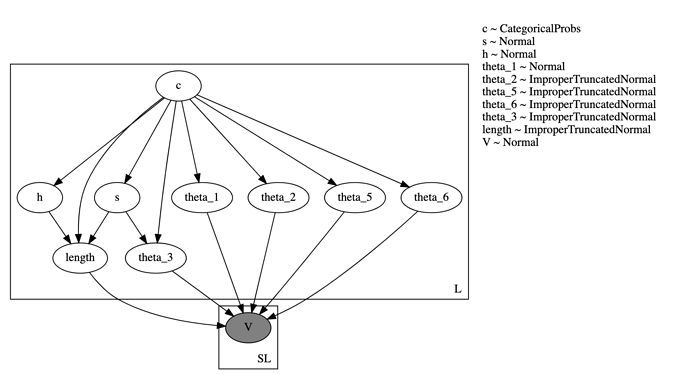
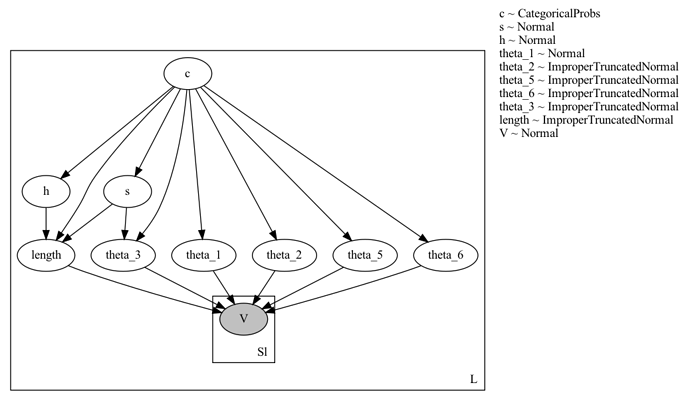
Model Description: I have a dataset of L objects. For each object, I sample a discrete variable c and eight continuous variables – s , h and six parameters theta_i which determine an analytical function (defined by the method dst). This function is fit to observed data points, one fit per object. Ideally the fitting code should be in a nested plate, but since the number of observed data points for each object maybe different, I have instead flattened out all the data points as a single vector V and do the fitting in a separate plate. The total number of data points is SL. The code and graphical model are at the end of this post.
When testing on a dataset of L ~ 3000, SL ~ 21000 , it takes ~ 5 minutes to run 4 chains with 1000 iterations each (500 warmup, 500 sample) on a machine with 8-core CPU and 32 GB ram. However, when testing on a larger dataset of L ~ 3e5 SL ~ 4e6, it takes ~1.5 days.
I am seeking advice on how to speed-up the sampling. I suspect that a large vectors of million data points is causing performance problems for numpyro. Is there any way to tell the sampler to run things in batches of smaller size?
The code. The distribution ImproperTruncatedNormal is just a Normal distribution with positive support as suggested in this thread.
class ImproperTruncatedNormal(dist.Normal):
support = dist.constraints.positive
funsor.distribution.make_dist(ImproperTruncatedNormal, param_names=("loc", "scale"))
def dst(theta, time):
return theta[..., 0] + 0.5*theta[..., 1] * (
jnp.tanh(theta[..., 4] * (time - theta[..., 2])) -
jnp.tanh(theta[..., 5] * (time - theta[..., 3]))
)
def my_model(V_obs, t, index_mapping, L, pi, theta_mean, theta_std, s_line_fit_params, h_line_fit_params, s_prior, h_prior, sigma, SL):
with numpyro.plate("L", L):
c = numpyro.sample("c", dist.Categorical(jnp.array(pi)), infer={'enumerate': 'parallel'})
s = numpyro.sample("s", dist.Normal(loc=jnp.array(s_prior[c, 0]), scale=jnp.array(s_prior[c, 1])))
h = numpyro.sample("h", dist.Normal(loc=jnp.array(h_prior[c, 0]), scale=jnp.array(h_prior[c, 1])))
theta_1 = numpyro.sample("theta_1", dist.Normal(loc=jnp.array(theta_mean[c, 0]), scale=jnp.array(theta_std[c, 0])))
theta_2 = numpyro.sample("theta_2", ImproperTruncatedNormal(loc=jnp.array(theta_mean[c, 1]), scale=jnp.array(theta_std[c, 1])))
theta_5 = numpyro.sample("theta_5", ImproperTruncatedNormal(loc=jnp.array(theta_mean[c, 2]), scale=jnp.array(theta_std[c, 4])))
theta_6 = numpyro.sample("theta_6", ImproperTruncatedNormal(loc=jnp.array(theta_mean[c, 3]), scale=jnp.array(theta_std[c, 5])))
gamma_3 = jnp.array(s_line_fit_params[c, 0] + s * s_line_fit_params[c, 1])
gamma_4 = jnp.array(h_line_fit_params[c, 0] + h * h_line_fit_params[c, 1])
gamma_length = gamma_4 - gamma_3
sigma_gamma_length = jnp.sqrt(theta_std[c, 3]**2 - theta_std[c, 2]**2)
theta_3 = numpyro.sample("theta_3", ImproperTruncatedNormal(loc=gamma_3, scale=theta_std[c, 2]))
length = numpyro.sample("length", ImproperTruncatedNormal(loc=gamma_length, scale=sigma_gamma_length))
theta_4 = numpyro.deterministic("theta_4", theta_3 + length)
theta = numpyro.deterministic("theta", jnp.stack([theta_1, theta_2, theta_3, theta_4, theta_5, theta_6], axis=-1))
with numpyro.plate("SL", SL):
v_t = dst(theta[..., index_mapping, :], t)
V = numpyro.sample("V", dist.Normal(v_t, sigma), obs=V_obs)
The graphical model