When learning guide function params in SVI, the shape of the param for the discrete latent variable is different than expected.
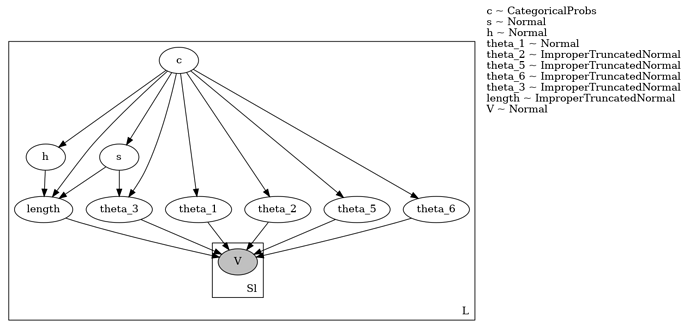
I have a numpyro model that I’m trying to move over from MCMC to SVI. The model is described in this post, the graphical model is shown below.
For SVI, I’m using TraceGraph_ELBO and defining a custom guide function as suggested in this post in the same thread. The guide function is below, along with the code used to run SVI.
def ci_guide(data, times, data_mask, L, pi, theta_mean, theta_std, s_line_fit_params, h_line_fit_params, s_prior, h_prior, sigma, Sl):
with numpyro.plate("L", L):
c_pi = numpyro.param("c_pi_q", pi, constraint=constraints.simplex, event_dim=2)
c = numpyro.sample("c", dist.Categorical(jnp.array(c_pi)), infer={'enumerate': 'parallel'})
s_mean = numpyro.param("s_mean_q", s_prior[c, 0])
s_std = numpyro.param("s_std_q", s_prior[c, 1], constraint=constraints.positive)
s = numpyro.sample("s", dist.Normal(loc=s_mean, scale=s_std))
h_mean = numpyro.param("h_mean_q", h_prior[c, 0])
h_std = numpyro.param("h_std_q", h_prior[c, 1], constraint=constraints.positive)
h = numpyro.sample("h", dist.Normal(loc=h_mean, scale=h_std))
theta_1_mean = numpyro.param("theta_1_mean_q", theta_mean[c, 0])
theta_1_std = numpyro.param("theta_1_std_q", theta_std[c, 0], constraint=constraints.positive)
theta_1 = numpyro.sample("theta_1", dist.Normal(loc=theta_1_mean, scale=theta_1_std))
theta_2_mean = numpyro.param("theta_2_mean_q", theta_mean[c, 1], constraint=constraints.positive)
theta_2_std = numpyro.param("theta_2_std_q", theta_std[c, 1], constraint=constraints.positive)
theta_2 = numpyro.sample("theta_2", ImproperTruncatedNormal(loc=theta_2_mean, scale=theta_2_std))
theta_5_mean = numpyro.param("theta_5_mean_q", theta_mean[c, 2], constraint=constraints.positive)
theta_5_std = numpyro.param("theta5_std_q", theta_std[c, 4], constraint=constraints.positive)
theta_5 = numpyro.sample("theta_5", ImproperTruncatedNormal(loc=theta_5_mean, scale=theta_5_std))
theta_6_mean = numpyro.param("theta_6_mean_q", theta_mean[c, 3], constraint=constraints.positive)
theta_6_std = numpyro.param("theta_6_std_q", theta_std[c, 5], constraint=constraints.positive)
theta_6 = numpyro.sample("theta_6", ImproperTruncatedNormal(loc=theta_6_mean, scale=theta_6_std))
gamma_3 = s_line_fit_params[c, 0] + s * s_line_fit_params[c, 1]
gamma_4 = h_line_fit_params[c, 0] + h * h_line_fit_params[c, 1]
gamma_length = gamma_4 - gamma_3
sigma_gamma_length = jnp.sqrt(theta_std[c, 3]**2 - theta_std[c, 2]**2)
theta_3_mean = numpyro.param("theta_3_mean_q", gamma_3, constraint=constraints.positive)
theta_3_std = numpyro.param("theta_3_std_q", theta_std[c, 2], constraint=constraints.positive)
theta_3 = numpyro.sample("theta_3", ImproperTruncatedNormal(loc=theta_3_mean, scale=theta_3_std))
length_mean = numpyro.param("length_mean_q", gamma_length, constraint=constraints.positive)
length_std = numpyro.param("length_std_q", sigma_gamma_length, constraint=constraints.positive)
length = numpyro.sample("length", ImproperTruncatedNormal(loc=length_mean, scale=length_std))
optimizer = numpyro.optim.Adam(step_size=0.0005)
svi = infer.SVI(my_model, ci_guide, optimizer, loss=infer.TraceGraph_ELBO(num_particles=10))
svi_result = svi.run(jrng_key, 2000, *args)
#L = 3000, K = 2, c_pi should have shape (2, 3000), instead has shape (2)
svi_result.params
'''
{'c_pi_q': DeviceArray([0.30955595, 0.69044405], dtype=float32),
'h_mean_q': DeviceArray([298.2612 , 287.16235, 287.19156, ..., 288.3742 , 298.34912,
298.35825], dtype=float32),
...
}
'''
In particular, I expect the parameter c_pi for the discrete latent variable c to be of shape (L, K) where K is the shape of the vector pi. Instead I get c_pi to just be a vector of shape K. All the remaining parameters are vectors of length L as expected. I tried adding event_dim=2 as per this post, but that didn’t change anything.
I’m unsure how to move forward, so any help would be appreciated.