I’m trying to implement a Bayesian neural network comprising anywhere from zero to two dense layers. In the the simplest case, with zero hidden layers, it’s essentially a linear regression model. The problem is that the model does not fit the training data well. From running a regular linear regression model I know that the bias (intercept) should be around -600, but my model (run without any dense layers) does not get anywhere close. It seems to me that the model gets stuck in a local minimum and can’t get out.
The model itself is loosely based on this blog post: Making Your Neural Network Say “I Don’t Know” — Bayesian NNs using Pyro and PyTorch | by Paras Chopra | Towards Data Science
I’ve read the related post High loss in Ordinary linear regression - #2 by dave but was not able to resolve the issue.
Help would be much appreciated!
class NN(nn.Module):
'''
The regression network.
'''
def __init__(self, input_size, hidden1_size, hidden2_size, dropout_rate):
super(NN, self).__init__()
self.input_size = input_size
self.hidden1_size = hidden1_size
self.hidden2_size = hidden2_size
self.dropout = nn.Dropout(p=dropout_rate)
if self.hidden1_size is None and self.hidden2_size is None:
self.out = nn.Linear(self.input_size, 1)
elif self.hidden1_size is not None and self.hidden2_size is None:
self.fc1 = nn.Linear(self.input_size, self.hidden1_size)
self.out = nn.Linear(self.hidden1_size, 1)
elif self.hidden1_size is None and self.hidden2_size is not None:
raise ValueError
elif self.hidden1_size is not None and self.hidden2_size is not None:
self.fc1 = nn.Linear(self.input_size, self.hidden1_size)
self.fc2 = nn.Linear(self.hidden1_size, self.hidden2_size)
self.out = nn.Linear(self.hidden2_size, 1)
def forward(self, x):
if self.hidden1_size is None and self.hidden2_size is None:
x = self.out(x)
elif self.hidden1_size is not None and self.hidden2_size is None:
x = self.dropout(F.leaky_relu(self.fc1(x)))
x = self.out(x)
elif self.hidden1_size is not None and self.hidden2_size is not None:
x = self.dropout(F.leaky_relu(self.fc1(x)))
x = self.dropout(F.leaky_relu(self.fc2(x)))
x = self.out(x)
return x
class BayesianRegressor(nn.Module):
'''
This PyTorch Module encapsulates the model as well as the
variational distribution (the guide) for the Regression Model
'''
def __init__(self, input_size, hidden1_size, hidden2_size, dropout_rate):
super().__init__()
self.df = 100
if hidden1_size is None and hidden2_size is None:
self.num_hidden = 0
elif hidden1_size is not None and hidden2_size is None:
self.num_hidden = 1
elif hidden1_size is None and hidden2_size is not None:
raise ValueError
elif hidden1_size is not None and hidden2_size is not None:
self.num_hidden = 2
# the network
self.net = NN(input_size,
hidden1_size,
hidden2_size,
dropout_rate)
self.softplus = torch.nn.Softplus()
self.DEVICE = 'cpu'
def model(self, X, y=None, importance=None):
# set all weights to 1.0 if not provided
if importance is None:
importance = torch.ones(X.shape[0])
# set prior for first fully connected layer
if self.num_hidden in [1, 2]:
fc1w_prior = Normal(loc=torch.zeros_like(self.net.fc1.weight),
scale=torch.ones_like(self.net.fc1.weight))
fc1b_prior = Normal(loc=torch.zeros_like(self.net.fc1.bias),
scale=torch.ones_like(self.net.fc1.bias))
# set priopr for second fully connected layer
if self.num_hidden == 2:
fc2w_prior = Normal(loc=torch.zeros_like(self.net.fc2.weight),
scale=torch.ones_like(self.net.fc2.weight))
fc2b_prior = Normal(loc=torch.zeros_like(self.net.fc2.bias),
scale=torch.ones_like(self.net.fc2.bias))
# set the prior for the final layer
outw_prior = Normal(loc=torch.zeros_like(self.net.out.weight),
scale=torch.ones_like(self.net.out.weight))
outb_prior = Normal(loc=torch.zeros_like(self.net.out.bias),
scale=torch.ones_like(self.net.out.bias))
if self.num_hidden == 0:
priors = {'out.weight': outw_prior, 'out.bias': outb_prior}
elif self.num_hidden == 1:
priors = {'fc1.weight': fc1w_prior, 'fc1.bias': fc1b_prior,
'out.weight': outw_prior, 'out.bias': outb_prior}
elif self.num_hidden == 2:
priors = {'fc1.weight': fc1w_prior, 'fc1.bias': fc1b_prior,
'fc2.weight': fc2w_prior, 'fc2.bias': fc2b_prior,
'out.weight': outw_prior, 'out.bias': outb_prior}
# lift module parameters to random variables sampled from the priors
lifted_module = pyro.random_module("module", self.net, priors)
# sample a regressor (which also samples w and b)
lifted_reg_model = lifted_module()
# make the prediction
y_hat = lifted_reg_model(X)
# sample the noise magnitude
sigma = pyro.sample('sigma', dist.LogNormal(loc=0., scale=1.))
with pyro.poutine.scale(scale=importance):
pyro.sample("obs", StudentT(df=self.df, loc=y_hat, scale=sigma), obs=y)
return y_hat
def guide(self, X, y=None, importance=None):
# First layer weight distribution priors
if self.num_hidden in [1, 2]:
fc1w_mu_param = pyro.param("fc1w_mu_param", torch.randn_like(self.net.fc1.weight))
fc1w_sigma_param = self.softplus(pyro.param("fc1w_sigma_param", torch.randn_like(self.net.fc1.weight)))
fc1w_prior = Normal(loc=fc1w_mu_param,
scale=fc1w_sigma_param)
# First layer bias distribution priors
fc1b_mu_param = pyro.param("fc1b_mu_param", torch.randn_like(self.net.fc1.bias))
fc1b_sigma_param = self.softplus(pyro.param("fc1b_sigma_param", torch.randn_like(self.net.fc1.bias)))
fc1b_prior = Normal(loc=fc1b_mu_param,
scale=fc1b_sigma_param)
if self.num_hidden == 2:
# Second layer weight distribution priors
fc2w_mu_param = pyro.param("fc2w_mu_param", torch.randn_like(self.net.fc2.weight))
fc2w_sigma_param = self.softplus(pyro.param("fc2w_sigma_param", torch.randn_like(self.net.fc2.weight)))
fc2w_prior = Normal(loc=fc2w_mu_param,
scale=fc2w_sigma_param)
# Second layer bias distribution priors
fc2b_mu_param = pyro.param("fc2b_mu_param", torch.randn_like(self.net.fc2.bias))
fc2b_sigma_param = self.softplus(pyro.param("fc2b_sigma_param", torch.randn_like(self.net.fc2.bias)))
fc2b_prior = Normal(loc=fc2b_mu_param,
scale=fc2b_sigma_param)
# Output layer weight distribution priors
outw_mu_param = pyro.param("outw_mu_param", torch.randn_like(self.net.out.weight))
outw_sigma_param = self.softplus(pyro.param("outw_sigma_param", torch.randn_like(self.net.out.weight)))
outw_prior = Normal(loc=outw_mu_param,
scale=outw_sigma_param)
# Output layer bias distribution priors
outb_mu_param = pyro.param("outb_mu_param", torch.randn_like(self.net.out.bias))
outb_sigma_param = self.softplus(pyro.param("outb_sigma_param", torch.randn_like(self.net.out.bias)))
outb_prior = Normal(loc=outb_mu_param,
scale=outb_sigma_param)
if self.num_hidden == 0:
priors = {'out.weight': outw_prior, 'out.bias': outb_prior}
elif self.num_hidden == 1:
priors = {'fc1.weight': fc1w_prior, 'fc1.bias': fc1b_prior,
'out.weight': outw_prior, 'out.bias': outb_prior}
elif self.num_hidden == 2:
priors = {'fc1.weight': fc1w_prior, 'fc1.bias': fc1b_prior,
'fc2.weight': fc2w_prior, 'fc2.bias': fc2b_prior,
'out.weight': outw_prior, 'out.bias': outb_prior}
# fit the noise
sigma_mu_param = pyro.param("sigma_mu_param", tensor(0.0))
sigma_sigma_param = self.softplus(pyro.param("sigma_sigma_param", tensor(1.0)))
sigma = pyro.sample('sigma', dist.LogNormal(loc=sigma_mu_param,
scale=sigma_sigma_param))
lifted_module = pyro.random_module("module", self.net, priors)
return lifted_module()
def fit(self, X, y, importance=None, lr=0.01, steps=5000):
"""Fit the model to data.
"""
self.steps = steps
# put the net into training mode (i.e. turn on drop-out if applicable)
self.net.train()
# Clear all the parameter store
pyro.clear_param_store()
# Define the SVI algorithm.
my_svi = SVI(model=self.model,
guide=self.guide,
optim=Adam({"lr": lr}),
loss=Trace_ELBO())
self.train_losses = []
for i in range(self.steps):
loss = my_svi.step(X, y, importance) / X.shape[0]
self.train_losses.append(loss)
if (i % 100 == 0):
print(f'iter: {i}, loss: {round(loss, 2)}', end="\r")
def predict(self, X, num_models=50, return_std=False):
"""Predict new samples
"""
self.net.eval()
predictive = Predictive(model=regressor.model,
guide=regressor.guide,
num_samples=800,
return_sites=("obs",))
samples = predictive(X)
y_hat = samples['obs'].mean(axis=0).detach().numpy()
sigma = samples['obs'].std(axis=0).detach().numpy()
if return_std is True:
return y_hat, sigma
else:
return y_hat
Preparing the data:
# dataset from scikit learn
data = sklearn.datasets.load_boston()
Xs = torch.tensor(data['data']).float()
ys = torch.tensor(data['target']).float()
# normalize input
Xs_norm = (Xs - Xs.mean()) / np.sqrt(Xs.var())
# split
Xs_train, Xs_test, ys_train, ys_test, = train_test_split(Xs_norm, ys,
shuffle=True,
test_size=0.2,
random_state=4321)
Xs_train.shape, Xs_test.shape, len(ys_train), len(ys_test)
Training:
%%time
steps = 5000
regressor = BayesianRegressor(input_size=Xs_train.shape[1],
hidden1_size=None,
hidden2_size=None,
dropout_rate=0.0)
regressor.fit(X=Xs_train,
y=ys_train,
steps=steps,
importance=None,
lr=0.01)
The bias term is no where near -600
regressor.net.out.bias
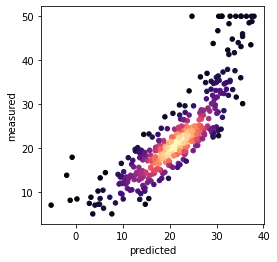
Predicting on the training data, just to show that the model has not fit it:
y_hat, sigma = regressor.predict(Xs_train,
num_models=800,
return_std=True)
x = y_hat.flatten()
y = ys_train
# Calculate the point density
xy = np.vstack([x,y])
z = scipy.gaussian_kde(xy)(xy)
fig, ax = plt.subplots(figsize=(4,4))
ax.scatter(x, y, c=z, s=20, cmap='magma')
plt.xlabel('predicted')
plt.ylabel('measured')
plt.show()