Hi!
I decided to try out Pyro/Numpyro to test the latest normalizing flow algorithms on my astrophysics model. The model is simple, consisting of only 7 parameters, but the posterior is usually multimodal (in a predictable way).
Here’s an outline of the model
def model(t, F, Ferr):
ln_DeltaF = numpyro.sample("ln_DeltaF", dist.Normal(4., 4.))
DeltaF = jnp.exp(ln_DeltaF)
ln_Fbase = numpyro.sample("ln_Fbase", dist.Normal(2., 4.))
Fbase = jnp.exp(ln_Fbase)
t0 = numpyro.sample("t0", dist.Normal(ca.utils.estimate_t0(event), 20.))
ln_tE = numpyro.sample("ln_tE", dist.Normal(3., 6.))
tE = jnp.exp(ln_tE)
u0 = numpyro.sample("u0", dist.Normal(0., 1.))
piEE = numpyro.sample("piEE", dist.Normal(0., .5))
piEN = numpyro.sample("piEN", dist.Normal(0., .5))
# Trajectory
zeta_e_t = jnp.interp(t, points, zeta_e)
zeta_n_t = jnp.interp(t, points, zeta_n)
zeta_e_t0 = jnp.interp(t0, points, zeta_e)
zeta_n_t0 = jnp.interp(t0, points, zeta_n)
zeta_e_dot_t0 = jnp.interp(t0, points, zeta_e_dot)
zeta_n_dot_t0 = jnp.interp(t0, points, zeta_n_dot)
delta_zeta_e = zeta_e_t - zeta_e_t0 - (t - t0) * zeta_e_dot_t0
delta_zeta_n = zeta_n_t - zeta_n_t0 - (t - t0) * zeta_n_dot_t0
u_per = u0 + piEN * delta_zeta_e - piEE * delta_zeta_n
u_par = (
(t - t0) / tE + piEE * delta_zeta_e + piEN * delta_zeta_n
)
u = jnp.sqrt(u_per**2 + u_par**2)
# Magnification
A_u = (u ** 2 + 2) / (u * jnp.sqrt(u ** 2 + 4))
A_u0 = (u0 ** 2 + 2) / (jnp.abs(u0) * jnp.sqrt(u0 ** 2 + 4))
A = (A_u - 1) / (A_u0 - 1)
# Predicted flux
F_pred = DeltaF*A + Fbase
ln_c = numpyro.sample("ln_c", dist.Exponential(1/2.))
numpyro.sample("data_dist", dist.Normal(F_pred, jnp.exp(ln_c) * Ferr), obs=F)
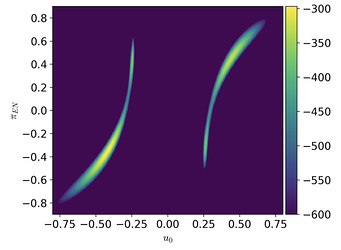
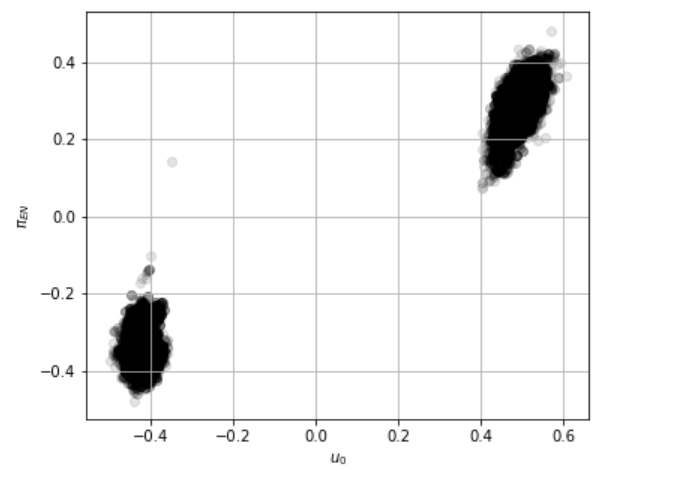
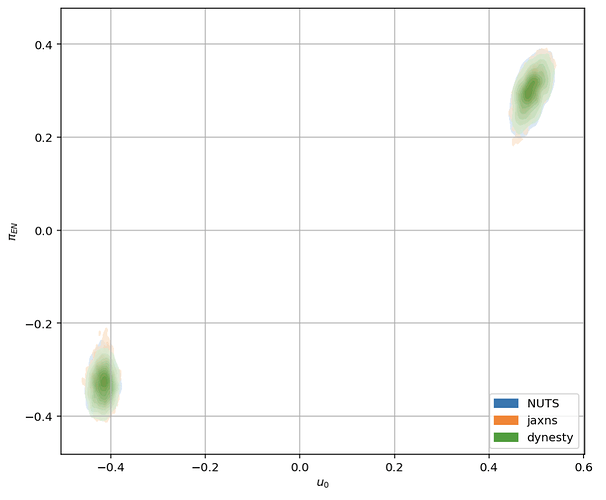
When I sample the model with NUTS the sampler finds only one of the modes because the posterior is multimodal in the u_0 parameter. Here’s what the likelihood surface looks like when evaluated on a (u0, piEN) grid while keeping all the other parameters fixed:
Ideally, I’d like to be able to train a guide which finds both modes and pass the guide to HMC for use in NeuTra HMC.
Based on this example, I thought the AutoBNAFNormal autoguide might be able to catch both modes, however, after testing various combinations of input parameters I find that it always gets stuck in one of the modes. Even when I fix all of the parameters and fit for the parameters in a 2D subspace of the full parameter space, it fails to find both modes except for a few very particular combinations of the learning rate and input parameters to AutoBNAFNormal.
My questions are the following:
- Is there a better choice for a guide which would work well for this kind of problem?
- I’ve read a paper tackling a similar problem where they used a 20 block MAF and a mixture of Gaussians for the base distribution of the flow and they seem to be able to recover multiple modes. How can I implement this in Pyro? Can I change the base distribution for
AutoBNAFNormal? I’ve read the docs on constructing custom guides but I don’t really know where to start.
Thanks!