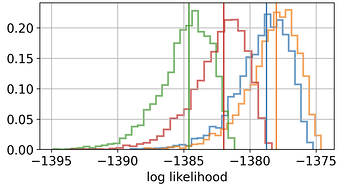
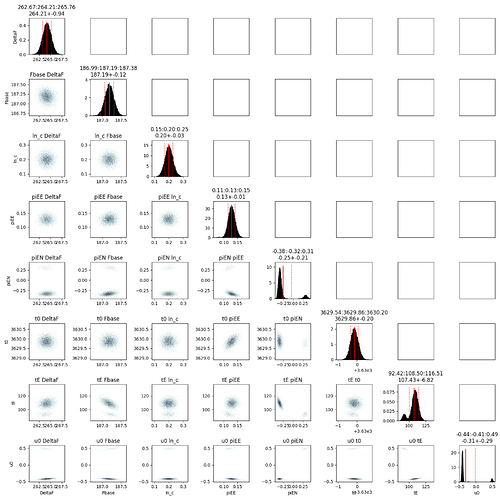
Hello,
I may be late (too) but I have just tried use the current Numpyro/NestedSampler for this quite interesting example. Following the discussion if I am right I have done this
from numpyro.contrib.nested_sampling import NestedSampler
ns = NestedSampler(model, num_live_points=2000, max_samples=2e5, depth=5, num_slices=3)
ns.run(random.PRNGKey(0), t, F, Ferr)
ns.print_summary()
I get first,
--------
# likelihood evals: 4000
# samples: 18355
# likelihood evals / sample: 0.2
--------
logZ=-3255.62 +- 0.058
ESS=2051
but then the code crash with the following last message
TypeError: percentile requires ndarray or scalar arguments, got <class 'list'> at position 1.
I think this is related to the following issue
samples = ns.get_samples(random.PRNGKey(1), 10000)
samples
one gets
{'ln_DeltaF': DeviceArray([-6.073782, -6.073782, -6.073782, ..., -6.073782, -6.073782,
-6.073782], dtype=float32),
'ln_Fbase': DeviceArray([5.2849283, 5.2849283, 5.2849283, ..., 5.2849283, 5.2849283,
5.2849283], dtype=float32),
'ln_c': DeviceArray([3.1351554, 3.1351554, 3.1351554, ..., 3.1351554, 3.1351554,
3.1351554], dtype=float32),
'ln_tE': DeviceArray([12.814183, 12.814183, 12.814183, ..., 12.814183, 12.814183,
12.814183], dtype=float32),
'piEE': DeviceArray([-0.26583126, -0.26583126, -0.26583126, ..., -0.26583126,
-0.26583126, -0.26583126], dtype=float32),
'piEN': DeviceArray([0.16695724, 0.16695724, 0.16695724, ..., 0.16695724,
0.16695724, 0.16695724], dtype=float32),
't0': DeviceArray([3578.5566, 3578.5566, 3578.5566, ..., 3578.5566, 3578.5566,
3578.5566], dtype=float32),
'u0': DeviceArray([0.1378763, 0.1378763, 0.1378763, ..., 0.1378763, 0.1378763,
0.1378763], dtype=float32)}
which is a unique point repeated all the time.
Is there something I missed to get NS working properly. (nb. NUTS/AutoBNAFNormal at least runs even if only 1 blob is found)
Thanks