Hi all,
I’ve been testing the new TraceGraph_ELBO implementation in numpyro for a sparse regression model and haven’t been able to get reasonable results. I’m unsure if this is due to variance in the gradients or if there is an aspect I am still missing.
The model I’ve implemented is the Sum of Single Effects model (ie SuSiE; see here). Even in a relatively low dimension (n = 400 and p = 10) the model seems unable to identify the variables with nonzero effect. I’ve provided the numpyro implementation alongside a simple implementation of the IBSS approach described in the above paper here.
Here is my model and guide:
# define the model
def model(X, y, l_dim) -> None:
n_dim, p_dim = jnp.shape(X)
# fixed prior prob for now
logits = jnp.ones(p_dim)
beta_prior_log_std = softplus_inv(jnp.ones(l_dim))
with numpyro.plate("susie_plate", l_dim):
gamma = numpyro.sample("gamma", dist.Multinomial(logits=logits))
beta_l = numpyro.sample("beta_l", dist.Normal(0.0, nn.softplus(beta_prior_log_std)))
# compose the categorical with sampled effects
beta = numpyro.deterministic("beta", beta_l @ gamma)
loc = X @ beta
std = numpyro.param("std", 0.5, constraint=constraints.positive)
with numpyro.plate("N", n_dim):
numpyro.sample("obs", dist.Normal(loc, std), obs=y)
return
def guide(X, y, l_dim) -> None:
n_dim, p_dim = jnp.shape(X)
# posterior for gamma
g_shape = (l_dim, p_dim)
gamma_logits = numpyro.param("gamma_post_logit", jnp.ones(g_shape))
# posterior for betas
b_shape = (p_dim, l_dim)
b_loc = numpyro.param("beta_loc", jnp.zeros(b_shape))
b_log_std = numpyro.param("beta_log_std", softplus_inv(jnp.ones(b_shape)))
with numpyro.plate("susie_plate", l_dim):
gamma = numpyro.sample("gamma", dist.Multinomial(logits=gamma_logits))
# average across individual posterior estimates
post_mu = numpyro.deterministic("post_mu", jnp.sum(gamma.T * b_loc, axis=0))
post_std = numpyro.deterministic("post_std", jnp.sum(gamma.T * nn.softplus(b_log_std), axis=0))
beta_l = numpyro.sample("beta_l", dist.Normal(post_mu, post_std))
return
And inference is being performed as:
scheduler = exponential_decay(args.learning_rate, 5000, 0.9)
adam = optim.Adam(scheduler)
svi = SVI(model, guide, adam, TraceGraph_ELBO())
# run inference
results = svi.run(
rng_key_run,
args.epochs,
X=X,
y=y,
l_dim=args.l_dim,
progress_bar=True,
stable_update=True,
)
The numpyro model results in a completely flat posterior and is unable to select the relevant variables, whereas the simple IBSS CAVI-style approach has no such troubles. I’ve tried different learning rates and decay with no luck. Similarly, increasing the number of particles didn’t seem to help much, which would suggest that variance in gradients isn’t necessarily the issue. Is there some other aspect I’m missing with the TraceGraph_ELBO capabilities?
EDIT: I updated the guide to explicitly average over individual posteriors and see improved performance, but not quite at what IBSS can capture.
EDIT2: Here is are renderings of the model and guide:
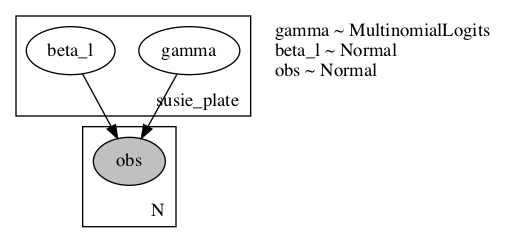
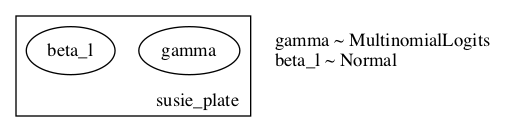
Given the explicit computation of beta_l moments from gamma I would have expected an arrow indicating the dependency in the posterior beta_l <- gamma, but I’m unsure if this is a result of the plate setup, or how rendering works when given a guide.