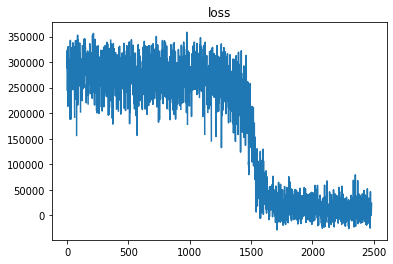
Hmm… I’m getting nans in my svi.step quite a bit, but not all the time.
Not sure how to trouble shoot this…
I tried decreasing the learning rate, increasing the batch size, using num_particles 10 to 100 in pyro.infer.Trace_ELBO(num_particles)
/usr/local/lib/python3.7/dist-packages/pyro/poutine/trace_struct.py:248: UserWarning: Encountered NaN: log_prob_sum at site 'quaternions'
"log_prob_sum at site '{}'".format(name),
/usr/local/lib/python3.7/dist-packages/pyro/infer/trace_elbo.py:158: UserWarning: Encountered NaN: loss
warn_if_nan(loss, "loss")
nan
def model(mini_batch,do_log=True):
"""
forward model with noise
point_cloud defined outside function
"""
size_mini_batch = mini_batch.shape[-1]
with pyro.plate('mini_batch',size_mini_batch):
quaternion_dist = dist.ProjectedNormal(concentration_cuda*q_loc_cuda)
quaternions = pyro.sample('quaternions',quaternion_dist)
rotations = pytorch3d.transforms.quaternion_to_matrix(quaternions)
clean_signal = do_forward_model(rotations, point_cloud) # backpropable
with pyro.plate('pixel_x',num_pix, dim=-2):
with pyro.plate('pixel_y',num_pix, dim=-3):
distrib = dist.Normal(clean_signal, sigma_noise)
pyro.sample("shot_noise", distrib, obs=mini_batch)
sim = distrib.sample()
return sim, quaternions
def guide(mini_batch,do_log=True): # the proposal distribution
"""
neural net will be trained on many particles to predict params of distribution
"""
pyro.module("net", net)
size_mini_batch = mini_batch.shape[-1]
with pyro.plate('mini_batch', size_mini_batch, dim=-1):
lam = net(mini_batch)
q_loc,log_concentration = lam[:,:4], lam[:,4]
q_unit_loc = q_loc / q_loc.norm(dim=1).reshape(-1,1)
concentration = torch.exp(log_concentration)
quaternion_dist = dist.ProjectedNormal(concentration.reshape(-1,1)*q_unit_loc)
quaternions = pyro.sample("quaternions", quaternion_dist)
return quaternions, lam
pyro.clear_param_store()
svi = pyro.infer.SVI(model=model,
guide=guide,
optim=pyro.optim.Adam({"lr": 1e-7}),
loss=pyro.infer.Trace_ELBO())