Thanks for the great package. I’m trying to sample from a mixture model, but am running into a (user) error. Below is a MWE which describes my problem. Hopefully it captures the problem I’m running into.
I basically took some code from the examples and tried to modify it in a sensible way for an application I’m working on. The model provided is a simplified one of the one I actually want to use. The problem with the model is with the control flow (if statements) in the below model. Note that data will be a vector of size N.
import torch
import pyro
import pyro.distributions as dist
from pyro.optim import Adam
from pyro.infer.autoguide import AutoDelta
from pyro.infer import SVI, TraceEnum_ELBO, config_enumerate
@config_enumerate
def model(data=None, size=None):
alpha = pyro.sample('alpha', dist.Dirichlet(torch.tensor([1.,2.,3.])))
with pyro.plate('b', size) as ind:
if data is not None:
with pyro.util.ignore_jit_warnings():
assert data.size == size
data = data[ind]
z = pyro.sample('z', dist.Categorical(alpha))
if z.item() == 0:
beta = pyro.sample('beta', dist.Normal(1,2))
elif z.item() == 1:
beta = pyro.sample('beta', dist.Normal(3,4))
else:
beta = pyro.sample('beta', dist.Normal(5,6))
data = pyro.sample('data', dist.Normal(beta**2, 0.01), obs=data)
return data
guide = AutoDelta(model)
optim = Adam({"lr": 0.01})
loss = TraceEnum_ELBO(max_plate_nesting=1)
svi = SVI(model, guide, optim, loss=loss, num_samples=1000)
def train(data, num_iterations=1000):
pyro.clear_param_store()
for j in range(num_iterations):
# calculate the loss and take a gradient step
loss = svi.step(data, data.size)
if j % 50 == 0:
print("[iteration %04d] loss: %.4f" % (j + 1, loss / len(data)))
x = model(size=100)
train(x)
This results in the error:
8 data = data[ind]
9 z = pyro.sample('z', dist.Categorical(alpha))
---> 10 if z.item() == 0:
11 beta = pyro.sample('beta', dist.Normal(1,2))
12 elif z.item() == 1:
ValueError: only one element tensors can be converted to Python scalars
As you can see, this fails since z is the same size as data which the .item() method doesn’t handle. (Edit: The above MWE actually fails on the model(size=100) call.) How should I write this to allow vectorized sampling? (Not sure if that is the right terminology.) Does a tutorial already exist for this that I’ve missed?
By the way, I was going to use the MixtureOfDiagNormals distribution, but this doesn’t handle D=1 data. Why is this the case?
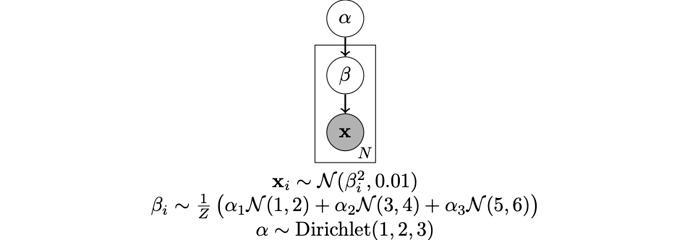
For context, in the above example I am trying to get the MAP estimates of the beta parameters in the following graphical model.
I’m providing this context because perhaps the whole setup is completely misguided.
FYI, I’m using pytorch version 1.2.0 and pyro version 0.4.1.
Thanks for the help. Apologies if this topic is addressed in a tutorial or in another thread or github issue.