My goal is to learn the relationship between a variable and the second parameter of a beta distribution. SVI fails with a ValueError stemming from NaNs in the MultivariateNormal guide.
Minimal Example:
# model definition
def model5(ac=None, c=None, N=None):
b0 = sample('b0', LogNormal(0., 0.1).to_event())
m = sample('m', Normal(1., 2.).to_event()) # slope
b = sample('b', Normal(1., 1.).to_event()) # intercept
b1_scale = sample('s', Exponential(1.))
N = N if N is not None else 20
with plate("data", N, dim=-1):
c = sample('c', Uniform(0., 1.), obs=c) # independent variable
b1_loc = m * c + b # linear function
b1 = sample('b1', LogNormal(b1_loc, b1_scale))
ac = sample('ac', BetaBinomial(b0, b1, total_count=252_000), obs=ac)
return ac, c
print(model5(N=5))
print_shapes(model5)
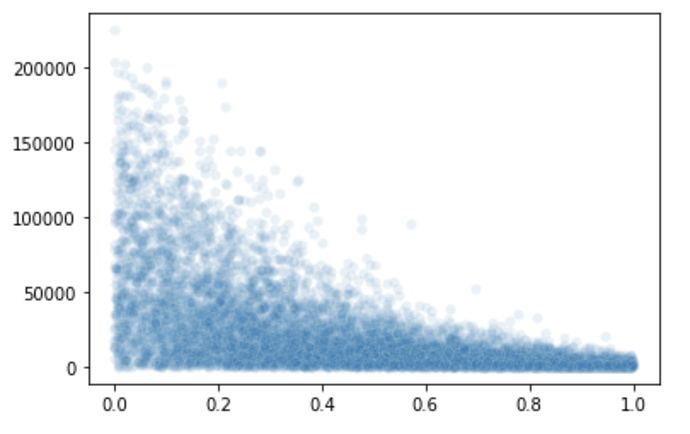
sns.scatterplot(x=res[1], y=res[0], alpha=0.1)
# condition and sample the model
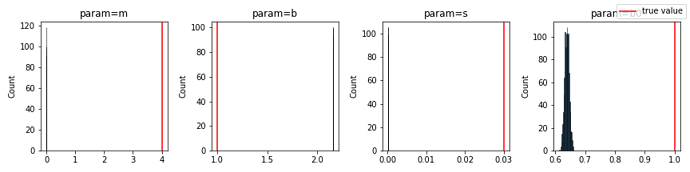
conditioned_model = pyro.poutine.condition(model5, data={'b': 1., 'm': 4})
res = conditioned_model(N=10000)
# SVI with multivariate normal guide
train_model(model5, obs=[res[0], res[1], 10000])
Outputs:
(tensor([ 1827., 45182., 186284., 250143., 248686.]),
tensor([0.2874, 0.8567, 0.2700, 0.5788, 0.7854]))
Trace Shapes:
Param Sites:
Sample Sites:
b0 dist |
value |
log_prob |
m dist |
value |
log_prob |
b dist |
value |
log_prob |
s dist |
value |
log_prob |
data dist |
value 20 |
log_prob |
c dist 20 |
value 20 |
log_prob 20 |
b1 dist 20 |
value 20 |
log_prob 20 |
ac dist 20 |
value 20 |
log_prob 20 |
Elbo loss: 120314.78125
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
File ~/Library/Caches/pypoetry/virtualenvs/statistical-rethinking-64KwZK9C-py3.9/lib/python3.9/site-packages/pyro/poutine/trace_messenger.py:174, in TraceHandler.__call__(self, *args, **kwargs)
173 try:
--> 174 ret = self.fn(*args, **kwargs)
175 except (ValueError, RuntimeError) as e:
...
...
...
ValueError: Expected parameter loc (Tensor of shape (10004,)) of distribution MultivariateNormal(loc: torch.Size([10004]), scale_tril: torch.Size([10004, 10004])) to satisfy the constraint IndependentConstraint(Real(), 1), but found invalid values:
tensor([ nan, nan, nan, ..., 2.3503, 2.1330, 1.2780],
grad_fn=<ExpandBackward0>)
Trace Shapes:
Param Sites:
AutoMultivariateNormal.scale 10004
AutoMultivariateNormal.scale_tril 10004 10004
AutoMultivariateNormal.loc 10004
Sample Sites:
Trace Shapes:
Param Sites:
AutoMultivariateNormal.scale 10004
AutoMultivariateNormal.scale_tril 10004 10004
AutoMultivariateNormal.loc 10004
Sample Sites:
I saw another forum post with a similar issue. The OP stated that setting all parameters to torch.float64 solved his/her issue. That does not work for me. Happy to provide that code if it helps.