Hi!
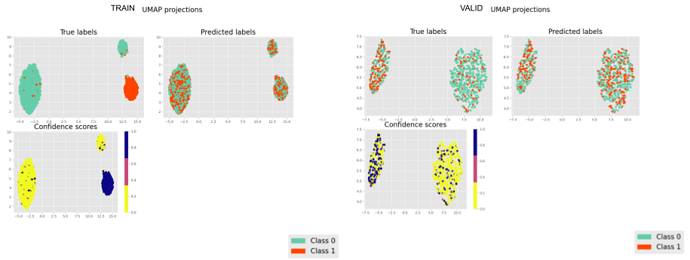
Inspired by the SSVAE (The Semi-Supervised VAE — Pyro Tutorials 1.8.4 documentation) I am building a supervised VAE (I have also unsupervised and semisupervised versions). I am currently puzzled because my latent representations cluster according to the binary classification, however, the NN classifier (no matter what architecture) is incapable of generating the right classifications.
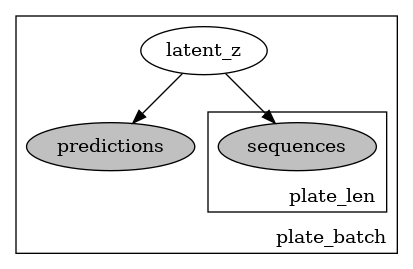
I would like some confirmation that the current architecture (here given over a dummy example) makes sense. I am worried about the correct use of the plates and the use of enumeration for discrete variables inference.
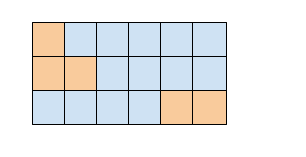
May you also please confirm that the masking over some positions across some positions in the sequence is valid? True means use this position to compute the likelihood, right? I am doubting everything at this point hehe
import torch
import torch.nn as nn
import pyro
import pyro.distributions as dist
from pyro.infer import config_enumerate,TraceEnum_ELBO,SVI
N = 50
L = 15
aa_types = 22
X = torch.randint(0,20,(N,L,aa_types)) #sequences
Y = torch.randint(0,1,(N,)) #labels
hidden_dim= 30
z_dim = 5
learning_type = "supervised"
class VAE_classifier(nn.Module):
def __init__(self):
super(VAE_classifier, self).__init__()
self.beta = 1 #no scaling right now
self.gru_hidden_dim = self.hidden_dim*2
self.aa_types = aa_types
self.max_len = L
self.z_dim = z_dim
self.num_classes = 2
self.learning_type = learning_type
self.encoder = RNN_encoder(self.aa_types,self.max_len,self.gru_hidden_dim,self.z_dim,self.device)
self.encoder_guide = RNN_encoder(self.aa_types,self.max_len,self.gru_hidden_dim,self.z_dim,self.device)
self.decoder = RNN_decoder(self.aa_types,self.seq_max_len,self.gru_hidden_dim,self.aa_types,self.z_dim ,self.device)
self.classifier_model = MLP(self.z_dim,self.max_len,self.hidden_dim,self.num_classes,self.device)
self.h_0_MODEL_encoder = nn.Parameter(torch.randn(self.gru_hidden_dim), requires_grad=True).to(self.device)
self.h_0_GUIDE = nn.Parameter(torch.randn(self.gru_hidden_dim), requires_grad=True).to(self.device)
self.h_0_MODEL_decoder = nn.Parameter(torch.randn(self.gru_hidden_dim), requires_grad=True).to(self.device)
self.logsoftmax = nn.LogSoftmax(dim=-1)
def model(self,batch_sequences,batch_mask,batch_true_labels,batch_confidence_scores):
pyro.module("vae_model", self)
batch_size = batch_sequences.shape[0]
assert batch_mask.shape == (batch_size,self.max_len)
confidence_mask = (batch_confidence_scores[..., None] > 0.7).any(-1)
confidence_mask_true = torch.ones_like(confidence_mask).bool()
assert confidence_mask.shape == (batch_size,)
init_h_0_encoder = self.h_0_MODEL_encoder.expand(self.encoder.num_layers * 2, batch_sequences.shape[0],self.gru_hidden_dim).contiguous()
z_mean,z_scale = self.encoder(batch_sequences,init_h_0_encoder)
with pyro.poutine.scale(scale=self.beta):
with pyro.plate("plate_latent", batch_size,device=self.device):
latent_space = pyro.sample("latent_z", dist.Normal(z_mean, z_scale).to_event(1)) # [n,z_dim]
latent_z_seq = latent_space.repeat(1, self.max_len).reshape(batch_size, self.max_len, self.z_dim)
with pyro.poutine.mask(mask=[confidence_mask if self.learning_type in ["semisupervised"] else confidence_mask_true][0]):
with pyro.plate("plate_class_seq",batch_size,dim=-1,device=self.device):
class_logits = self.classifier_model(latent_space)
class_logits = self.logsoftmax(class_logits)
assert class_logits.shape == (batch_size, self.num_classes)
if self.learning_type == "semisupervised":
pyro.sample("predictions", dist.Categorical(logits=class_logits).to_event(1).mask(confidence_mask),obs=batch_true_labels,infer={'enumerate': 'parallel'})
elif self.learning_type == "supervised":
pyro.sample("predictions", dist.Categorical(logits=class_logits).to_event(1),obs=batch_true_labels,infer={'enumerate': 'parallel'})
else:
pyro.sample("predictions", dist.Categorical(logits=class_logits).to_event(1))
init_h_0_decoder = self.h_0_MODEL_decoder.expand(self.decoder.num_layers * 2, batch_size,self.gru_hidden_dim).contiguous()
with pyro.poutine.mask(mask=batch_mask): ## DOUBT: I want to mask some positions in the sequence, is this enough? I checked the trace and seemed correct
with pyro.plate("data_len",self.max_len,device=self.device):
with pyro.plate("data", batch_size,device=self.device):
sequences_logits = self.decoder(latent_z_seq,init_h_0_decoder)
sequences_logits = self.logsoftmax(sequences_logits)
pyro.sample("sequences",dist.Categorical(logits=sequences_logits).mask(batch_mask),obs=batch_sequences) # DOUBT: Double masking redundant?
def guide(self, batch_sequences,batch_mask,batch_true_labels,batch_confidence_scores):
pyro.module("vae_guide", self)
batch_size = batch_sequences.shape[0]
confidence_mask = (batch_confidence_scores[..., None] < 0.7).any(-1) #now we try to predict those with a low confidence score
confidence_mask_true = torch.ones_like(confidence_mask).bool()
init_h_0 = self.h_0_GUIDE.expand(self.encoder_guide.num_layers * 2, batch_size,self.gru_hidden_dim).contiguous() # bidirectional
with pyro.poutine.scale(scale=self.beta):
with pyro.plate("plate_latent", batch_size,device=self.device): #dim = -2
z_mean, z_scale = self.encoder_guide(batch_sequences, init_h_0)
assert z_mean.shape == (batch_size, self.z_dim), "Wrong shape got {}".format(z_mean.shape)
assert z_scale.shape == (batch_size, self.z_dim), "Wrong shape got {}".format(z_scale.shape)
latent_space = pyro.sample("latent_z", dist.Normal(z_mean,z_scale).to_event(1)) # [z_dim,n]
if self.learning_type in ["semisupervised","unsupervised"]:
with pyro.poutine.mask(mask=[confidence_mask if self.learning_type in ["semisupervised"] else confidence_mask_true][0]):
with pyro.plate("plate_class_seq", batch_size, dim=-1, device=self.device):
class_logits = self.classifier_guide(latent_space)
class_logits = self.logsoftmax(class_logits)
assert class_logits.shape == (batch_size,self.num_classes)
if self.learning_type == "semisupervised":
pyro.sample("predictions", dist.Categorical(logits=class_logits).to_event(1).mask(confidence_mask), obs=batch_true_labels,infer={'enumerate': 'parallel'})
else: #unsupervised
pyro.sample("predictions", dist.Categorical(logits=class_logits).to_event(1))
guide = config_enumerate(VAE_classifier.guide)
optimizer = pyro.optim.ClippedAdam(dict())
svi = SVI(VAE_classifier.model, guide, optimizer, TraceEnum_ELBO(max_plate_nesting=1))
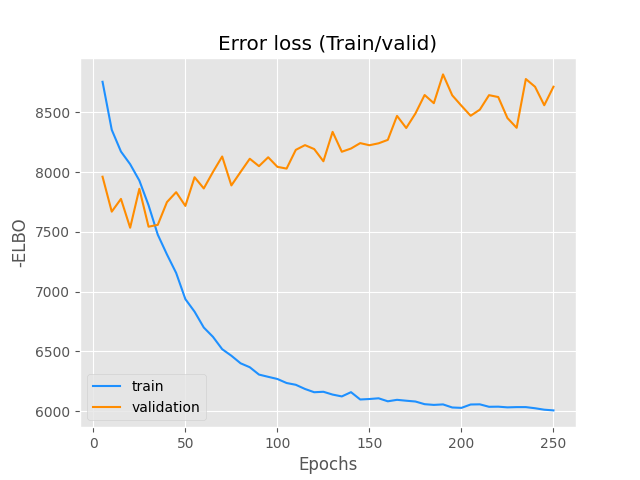
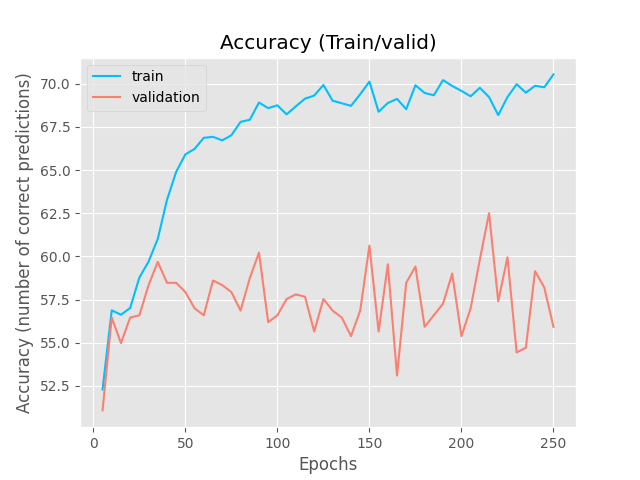
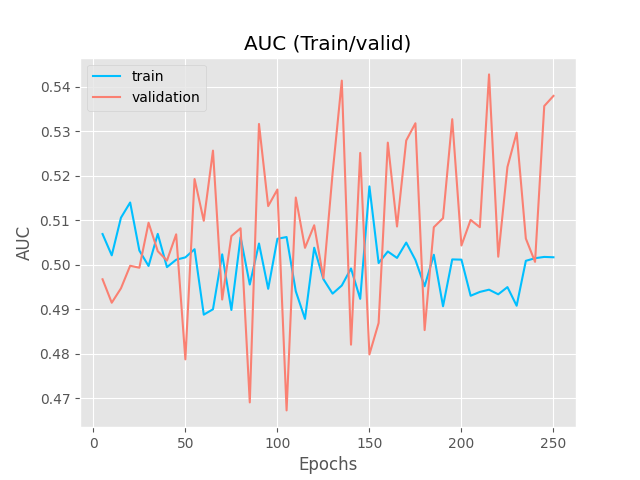
The encoder and decoders are just simple RNN + FCL . The NN architecture of the classifier is a simple MLP, since I tried more complex architectures and it would only predict one class (I also think it does not need it, given the nice clustering). I do not think is a hyperparameter issue either.
I want to confirm that the usage of config_enumerate, TraceEnum_ELBO, the plates and the masks makes sense, or what should I look into? Or should I use Trace_ELBO()?
Are the unsupervised and semisupervised modes making sense?
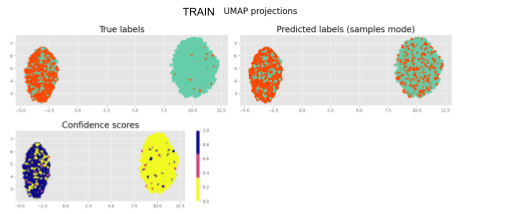
It is strange to me that the latent representations cluster perfectly and the NN classifier cannot figure out the right prediction ( I have checked my code for errors unrelated to the modelling and could not find bugs (for now)).
Note that the supervised model outperforms the unsupervised, therefore the values of the observed labels are being used to infer the latent random variables.
That is why I think there is something off with my approach to binary classification of the latent space,
Thanks in advance!!!