Dear Pyro Community,
I have tried making a simple Beta-Bernoulli latent feature model, where variables are drawn from different distributions depending on the particular draw from the Bernoulli distribution. I use a normal probabilistic forward model, but a guide with a neural network for the task.
Training seems to go well and I get a low ELBO at the end; unfortunately, it seems to infer that all values from the Bernoulli distribution are equal to 0, which is incorrect around 20% of the time for the synthesized example data I used.
The complete code is available on Google Colab, but I have included the model/guide below:
def model_guide_gen(is_guide, encoder: Encoder=None):
def mg(data):
D = len(data)
if is_guide:
pyro.module('encoder', encoder)
means_pos = pyro.sample('means_pos', dists.HalfNormal(scale=0.1 * data.new_ones(F)).independent(1))
means_neg = pyro.sample('means_neg', dists.Normal(loc=2.5 * data.new_ones(F), scale=1 * data.new_ones(F)).independent(1))
means = torch.stack((means_neg, means_pos))
sd_pos = 0.1 * data.new_ones(F)
sd_neg = data.new_ones(F)
sd = torch.stack((sd_neg, sd_pos))
if is_guide:
c1, c0 = encoder.forward(data)
else:
c1 = 0.1 * data.new_ones((D, F))
c0 = 0.1 * data.new_ones((D, F))
for d in pyro.irange('data', D):
r = pyro.sample('r_%d' % d, dists.Beta(c1[d], c0[d]).independent(1))
for f in pyro.irange('features_%d' % d, F):
b = pyro.sample('b_%d_%d' % (d, f), dists.Bernoulli(r[f])).long()
if not is_guide:
pyro.sample('data_obs_%d_%d' % (d, f), dists.Normal(loc=means[b, f], scale=sd[b, f]), obs=data[d,f])
return mg
Do you have an idea of what might be wrong?
Thank you very much in advance!
1 Like
A few observations from your model:
- Since you have a discrete latent, consider using
TraceEnum_ELBO instead which will have a much lower variance, as it will marginalize out the discrete b.
- That said, I am not sure if that will solve your problem. You have a mixture of samples from a half cauchy and uniform, and it will be tough to separate the two (just plot the distribution). It would be understandable if most data points got collapsed onto the normal with mean 2.5. The tutorial on gaussian mixture models may be of interest to you if you want to learn the cluster parameters instead of using a fixed one as you are doing here.
1 Like
Thank you for the suggestions!
I will try TraceEnum_ELBO instead to see if it helps.
Regarding, the difference between Half-Cauchy and Uniform distribution. I agree that it might be hard to see the difference for many points, but there should still be a big difference regarding probability around 0.
I have included the plot of the two distributions below:
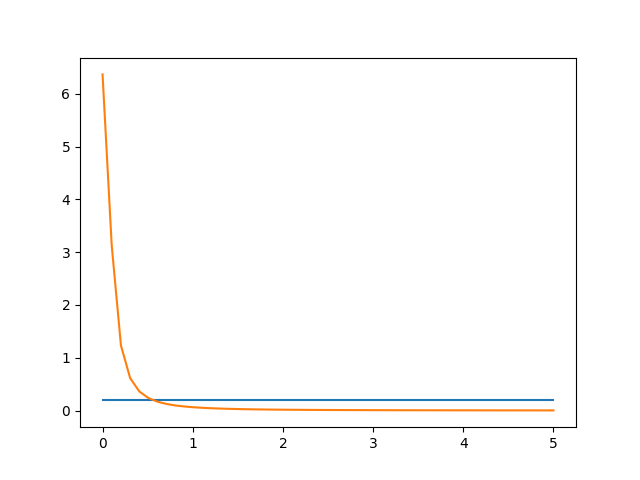
If I try a naïve comparison (as I did in the notebook), i.e.,
stupid_bs = (dists.HalfNormal(scale=0.1).log_prob(tdata) >= dists.Normal(loc=2.5, scale=1).log_prob(tdata)).detach().numpy()
I get at least only half of the 20% of 1s wrong, which is a bit better than the result I got by doing inference using my Pyro model. Could there be anything else that is wrong, e.g. my set-up of the neural network?
I think that since the points are not equally distributed (samples from the uniform are four times more likely), this plot makes the problem seem easier than it might be.
Some suggestions for debugging:
- I would suggest removing the for loops and vectorizing your model, which will make it run much faster.
- Maybe change the priors on c1, c0 from
0.1 to something like 1.0 that doesn’t force the model to choose either 0 or 1.
- Compare the log likelihood of your naive model by conditioning on both r (take 0.1 and 0.9 as per whether the b values are 0 or 1) and naive b, and compare it with the log likelihood of the learnt model. I will be very surprised if this is higher for the naive model.
1 Like
Thanks for the suggestions! I will make sure to try them out.
I appreciate all the help so far!
Using TraceEnum_ELBO did reduce variance, and vectorizing the data seemed to have provided a great boost to performance! I was unsure how to vectorize the inner-loop, but the performance has been good enough that I did not bother.
It does however seem that my naïve method has a higher log-probability than the inferred one, unless I am somehow miscalculating. Any other ideas?
Thanks again!
Please See:
My apologies for the late reply. Unless I’m mistaken, it seems like you are not accounting for the probability of the latent beta in your likelihood computation.
1 Like
No worries, I appreciate all the help!
I just used the point estimate for the expectation of the Beta distribution, since the variance was negligible for the inferred parameters (and so I believe it would only marginally change the likelihood).
It is hard for me to say where the issue might be without debugging further, for which I have some suggestions to begin with:
- Can we fix
means_pos to 0s and means_neg to 1s (EDIT: 2.5) to remove some stochasticity and simplify the problem?
- Can you try just training your encoder using binary cross entropy loss (you can generate some samples from a beta distribution and average the BCE loss), and see if you are able to recover your expected results? This might help you narrow down the issue (whether it’s somewhere in NN, or in SVI).
1 Like
Thank you very much!
I found the issue, was that I was conditioning my observations using a Normal distribution where the standard deviation was higher for the positive one, which made it much more unlikely than the negative one. This became more clear when I tried the first option you mentioned where I fixed the means.
1 Like
