Now that I have MAP prediction working thanks to help from this post I’m expanding my toy model closer to my actual application, but I have some followup questions
a is still Bernoulli and b is still a mixture of Bernoullis given a, and are very likely to take the value of a. But now instead of belonging to nested plates they’re in separate plates, and there is a new mapping, eg M = [0 0 1 0 1] that denotes which a_i dictates the mixture for each b_j. This is kind of unconventional I think but needed for further expansion towards my application (explained in the digression at the bottom).
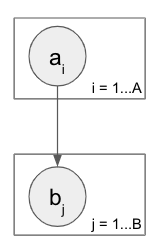
I’ve been able to get this working easily enough with sequential plates. The toy dataset has a = [0, 1] and the first five b belong to the first a and are thus 0, and vice versa for the second five b. With this setup I’m able to correctly predict a given b and also b given a.
MWE w/ sequential plates
import torch
import pyro
import pyro.distributions as dist
from pyro.infer import infer_discrete, config_enumerate
from pyro.ops.indexing import Vindex
import pyro.optim
from pyro import poutine
pyro.enable_validation(True)
num_b = 10
a_cpd = torch.tensor(0.9)
b_cpd = torch.tensor([.01, .99])
# first half of a and b are 0, second half are 1
data = {}
data['a'] = torch.tensor([0., 1.])
data['b'] = torch.cat((torch.zeros(num_b//2), torch.ones(num_b//2)))
data['map'] = torch.cat((torch.zeros(num_b//2), torch.ones(num_b//2)))
@config_enumerate
def model(a_obs=None, b_obs=None):
# sample a
a = []
for i in pyro.plate("plate_a", size=2):
a.append(pyro.sample(f'a_{i}',
dist.Bernoulli(a_cpd),
obs=a_obs[i] if a_obs is not None else None
))
# sample b
for i in pyro.plate("plate_b", size=num_b):
a_ind = int(data['map'][i])
a_cat = a[a_ind].long()
pyro.sample(f'b_{i}',
dist.Bernoulli(Vindex(b_cpd)[a_cat]),
obs=b_obs[i] if b_obs is not None else None
)
inferred_model = infer_discrete(model, temperature=0, first_available_dim=-2)
for target_var in ["a", "b"]:
kwargs = {"a_obs": data["a"].float()} if target_var == "b" else {"b_obs": data["b"].float()}
trace = poutine.trace(inferred_model).get_trace(**kwargs)
for key, val in sorted(trace.nodes.items()):
if key[0] == target_var:
print(key, val["value"])
The issue is that B could get very large, and the model itself will grow too, so the exponential runtime explosion for discrete enumeration with sequential plates becomes prohibitive. Given Restriction 2 for parallel plates used in discrete enumeration, I don’t expect to be able to parallelize the a_plate. But as far as I can tell I should be able to do so for the b_plate.
With a parallel b_plate I can get the prediction working for b given a, but can’t do it for predicting a given b. The issue comes in the call to model when a is enumerated, in which case I can’t figure out how to properly construct the mapped_cpd that compiles the appropriate cpd for each b based on its corresponding enumerated a.
Should it indeed be possible to parallelize the b_plate? If so, what would be the correct form of mapped_cpd to pass into the b sample site below when a is being enumerated? If not, is there an appropriate approximation method to use in place of this to avoid the runtime blowup?
M(non)WE with parallel `b_plate`
import torch
import pyro
import pyro.distributions as dist
from pyro.infer import infer_discrete, config_enumerate
from pyro.ops.indexing import Vindex
import pyro.optim
from pyro import poutine
pyro.enable_validation(True)
num_b = 10
a_cpd = torch.tensor(0.9)
b_cpd = torch.tensor([.01, .99])
# first half of a and b are 0, second half are 1
data = {}
data['a'] = torch.tensor([0., 1.])
data['b'] = torch.cat((torch.zeros(num_b//2), torch.ones(num_b//2)))
data['map'] = torch.cat((torch.zeros(num_b//2), torch.ones(num_b//2)))
@config_enumerate
def model(a_obs=None, b_obs=None):
# sample a
a = []
for i in pyro.plate("plate_a", size=3):
a.append(pyro.sample(f'a_{i}',
dist.Bernoulli(a_cpd),
obs=a_obs[i] if a_obs is not None else None
))
# sample b
with pyro.plate("plate_b", size=num_b):
a_ind = data['map']
if len(a[0].shape) > 0: # a is being enumerated
pass # THIS BRANCH IS THE ISSUE
else:
# pick out the outcome of the a associated with each b
a_val = torch.tensor([a[int(i)] for i in a_ind]).long()
mapped_cpd = Vindex(b_cpd)[a_val] # pick out the cpd val for each b given its a's outcome
pyro.sample('b',
dist.Bernoulli(mapped_cpd),
obs=b_obs
)
inferred_model = infer_discrete(model, temperature=0, first_available_dim=-2)
for target_var in ["b", "a"]:
kwargs = {"a_obs": data["a"].float()} if target_var == "b" else {"b_obs": data["b"].float()}
trace = poutine.trace(inferred_model).get_trace(**kwargs)
for key, val in sorted(trace.nodes.items()):
if key[0] == target_var:
print(key, val["value"])
Below is the digression regarding why I want to do this wonky mapping idea in the first place…
Digression
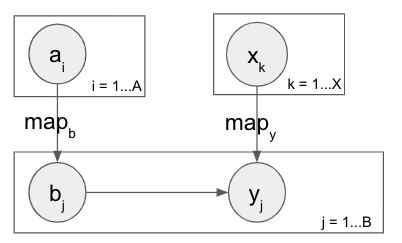
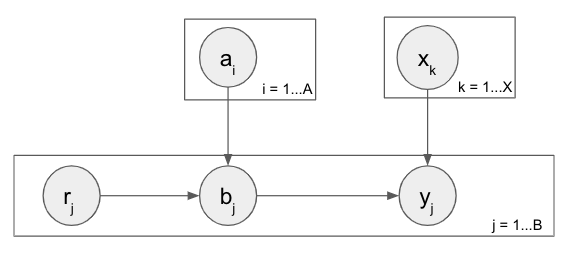
What I’m ultimately trying to do is predict machine/object failure in a processing environment. In the expanded model below, there’s some number of raw objects r that can be failing or healthy and enter a system, acted upon by machines of different types in multiple steps. In the each step there’s some number of machines (a and x) that can be failing or healthy, the raw objects are each assigned to one of the machines, and then b and y represent the health of the object after it’s been processed by the machine in that step. So the health of an object after step 1 depends on the health of the object before being passed into step 1 as well as the machine it ran through in step 1. And same thing for the object in step 2, which has a potentially different number of machines, and thus a completely different mapping from step 1.
Is this at all a sane approach to this sort of problem? Am I missing a much simpler approach?
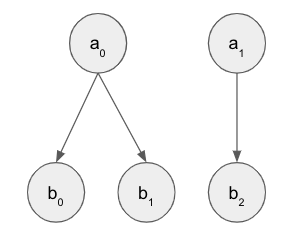
 . Thanks a ton to both of you for your help/ideas on this!
. Thanks a ton to both of you for your help/ideas on this!