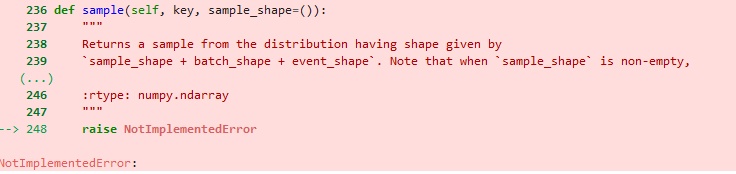
I am playing with some basic implementations of SVI in NumPyro, and having issues getting my guide’s to work for all but the most basic of dummy distributions. The simple example I’ve been working with is using SVI to recover a two-variable gaussian distribution. I have seen people use numpyro.factor() in other posts on this forum, so I know this should be a workable approach, but I am not getting consistent results. E.g. for the models:
def model_factor():
'''Gaussian distribution using factor()'''
x = numpyro.sample('x', dist.Uniform( -10 , 10))
y = numpyro.sample('y', dist.Uniform( -10 , 10))
log_fac = -1/2 * ( (x/sig1)**2 + (y/sig2)**2 )
log_fac -= jnp.log(sig1*sig2)
numpyro.factor('logfac', log_fac)
def model_nofactor():
'''Gaussian without using factor()'''
x = numpyro.sample('x', dist.Normal( 0 , sig1))
y = numpyro.sample('y', dist.Normal( 0 , sig2))
And the guides:
def guide_factor():
'''Gaussian guide using factor()'''
sig1_q = numpyro.param('sig1_q', 2.0, constraint = constraints.positive)
sig2_q = numpyro.param('sig2_q', 2.0, constraint = constraints.positive)
x = numpyro.sample('x', dist.Uniform( -10 , 10))
y = numpyro.sample('y', dist.Uniform( -10 , 10))
log_fac = -1/2 * ( (x/sig1)**2 + (y/sig2)**2 )
log_fac -= jnp.log(sig1*sig2)
numpyro.factor('log_fac', log_fac)
def guide_nofactor():
'''Gaussian guide without using factor()'''
sig1_q = numpyro.param('sig1_q', 1.0, constraint = constraints.positive)
sig2_q = numpyro.param('sig2_q', 1.0, constraint = constraints.positive)
x = numpyro.sample('x', dist.Normal( 0 , sig1_q))
y = numpyro.sample('y', dist.Normal( 0 , sig2_q))
I recover the correct results for sig_1 and sig_2 when using guide_nofactor, but not guide_factor, even though the probabilistic modelling is equivalent.
In case it ends up being relevant, I am calling them like:
svi_FF = SVI(model_factor, guide_factor, optimizer, loss=Trace_ELBO())
svi_FN = SVI(model_factor, guide_nofactor, optimizer, loss=Trace_ELBO())
svi_NF = SVI(model_nofactor, guide_factor, optimizer, loss=Trace_ELBO())
svi_NN = SVI(model_nofactor, guide_nofactor, optimizer, loss=Trace_ELBO())
#-------------------------------------------------------------------------
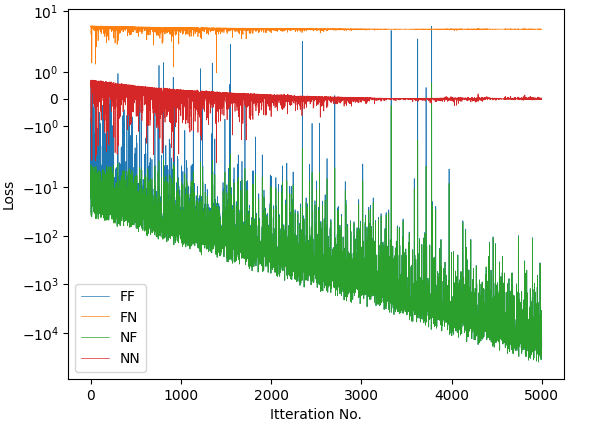
for svi, label in zip([svi_FF, svi_FN, svi_NF , svi_NN ], ["FF", "FN", "NF", "NN"]):
svi_result = svi.run(random.PRNGKey(1), 5000)
print(label)
for key, val in zip(svi_result.params.keys(), svi_result.params.values()):
print("%s:\t%0.3f" %(key,val) )
And am getting outputs that are consistent between models, but not guides. I’ve tried different tweaks to the log factor in guide_factor(), but similarly poor results.
FF
sig1_q: 0.715
sig2_q: 0.714
FN
sig1_q: 1.025
sig2_q: 1.994
NF
sig1_q: 0.715
sig2_q: 0.714
NN
sig1_q: 1.025
sig2_q: 1.994