Hello
I have been using numpyro for quite some time and find it very useful!
Recently I tried to implement a model which has 2 obs sites: 1) a fixed observed input and 2) a random variable.
A simplified version of the model is as below:
log (y_train) ~ N(a+b*log(h_train), e)
I have a set of y_train and h_train from which I want to learn the parameters a, b and e
Then I want to draw a new y_star conditional on a[j], b[j], e[j] and h_star. [j] represent each iteration in the mcmc chain. I am using NUTS sampler
Next I model the y_star using a set of x_inputs
y_star ~ Gamma(mu,sig)
mu = x_input * beta
sig = x_input * phi
The goal is to learn a, b, e, beta and phi all together.
I tried to used numpyro.param('a_j',a) to detach it from the log_prob calculation in the second observed statement but I realized that it only returns the initial_value, and not the value ‘a’ takes in each iteration.
My implementation is given below:
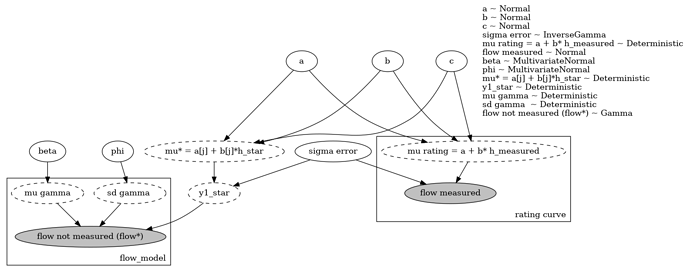
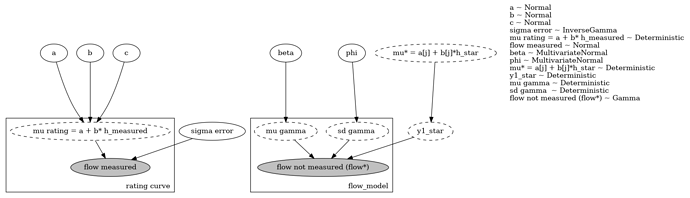
def full_model_single(y1_rating,h1_rating,x,h1_star):
n_train = len(y1_rating)
n = x.shape[1]
#Rating curve parameters
a = numpyro.sample('a', dist.Normal(0,n_train*10))
b = numpyro.sample('b', dist.Normal(0,n_train*10))
sigma2 = numpyro.sample('sigma error', dist.InverseGamma(1,2))
with numpyro.plate('rating curve',n_train):
mu = numpyro.deterministic('mu rating = a + b* h_measured', a + b*h1_rating)
numpyro.sample('flow measured', dist.Normal(mu, sigma2), obs=y1_rating)
# Fixing them to the values drawn
# But I realized this is wrong
a_j = numpyro.param('a_j', a)
b_j = numpyro.param('b_j', b)
sigma2_j = numpyro.param('sigma2_j', sigma2)
#Flow model parameters
beta = numpyro.sample('beta',dist.MultivariateNormal(jnp.zeros(3),covariance_matrix = n*jnp.eye(3)))
phi = numpyro.sample('phi',dist.MultivariateNormal(jnp.zeros(3),covariance_matrix = n*jnp.eye(3)))
mu_star = numpyro.deterministic('mu* = a[j] + b[j]*h_star', a_j + (b_j*h1_star) )
#Draw a y_star using the sampled a, b and sigma
y1_log_star = dist.Normal(mu_star ,sigma2_j).sample(rng_key_)
y1_star = numpyro.deterministic('y1_star', jnp.exp(y1_log_star))
with numpyro.plate('flow model',n):
mean = numpyro.deterministic('mu gamma', jnp.exp(x.T @ beta))
sd = numpyro.deterministic('sd gamma ', jnp.exp(x.T @ phi))
numpyro.sample("flow not measured (flow*)", dist.Gamma(mean**2/sd**2,rate=mean/sd**2), obs=y1_star)
This application is in hydrology where ‘h’ is measured height of flow at a station and ‘y’ is the volume of streamflow discharge.
Any help or leads would be very much appreciated!
Cheers
Rajitha