Hi, I’d like to ask for Your help in understading why my numpyro model is sampling very poorly (slow, lots of divergencies, stratospheric r_hat). The reason, why I feel somewhat entitled to being surprised by this poor sampling is that (what I think is) an equivalent model in pymc samples fine (same data, same priors, same model structure, and even using “numpyro” NUTS sampler in pymc). In other words I need help porting a model from pymc to numpyro.
Below I’m pasting my code, including dummy data generations (aligned with the model structure), defining both the pymc and numpyro versions of the model, and the attempts at sampling from both of them.
Few words about the model. It is a somewhat exotic 1D mixture. It’s made out of a) LogSkewNormal which is meant to describe majority of the data, and b) shifted Pareto distribution (ie LogExponential with two-parameter affine transformation on top) which is meant to soak up outliers which are expected to be rare and to lie to the left of the LogSkewNormal.
I’m genuinely invested in trying to fit this model to my real data at work, mostly for inferring parameters of the LogSkewNormal. The dummy data below is similar to one (out of many) of my datasets. And the priors below are fairly genuine: in particular I’m not assuming a priori that the components of the mixture are super well separated, despite the fact the it so happens in the dummy data below.
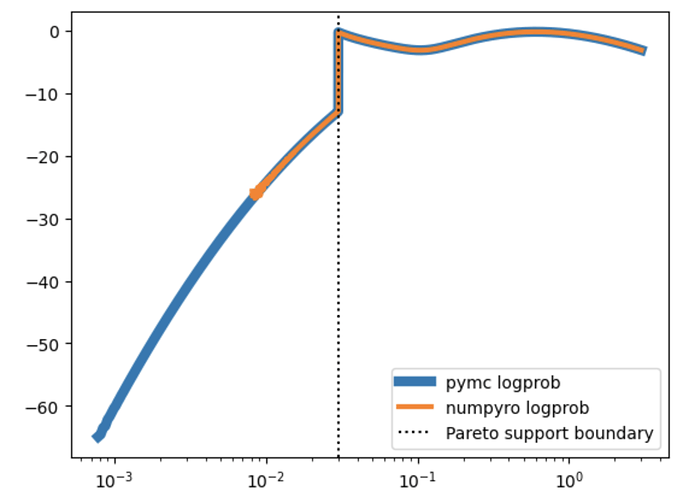
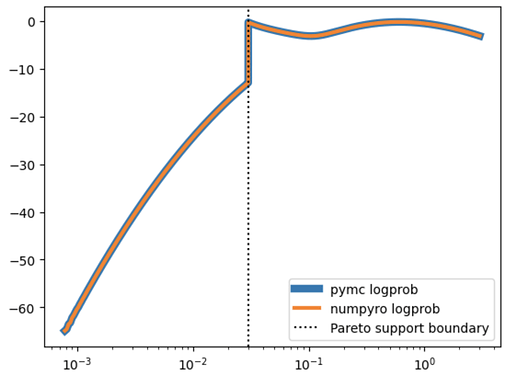
I’ve also repeated the exercise below but having only one component of the mixture at a time (ie. throwing the other component away from both data generation and the model). In such simple setting both pymc and numpyro models sample equally fine and produce compatible results (fairly correctly inferring the ground truth parameters). This makes me believe that my handmade implementations of LogSkewNormal and ShiftedPareto below are correct (and consistent between pymc and numpyro). That leaves me with the hypothesis that the problem lies somewhere in numpyro’s MixtureGeneral or my usage of it.
I would really appreciate any tips or discussion. Many thanks!
import numpy as np
import matplotlib.pyplot as plt
from scipy.stats import bernoulli, pareto, skewnorm
import pymc as pm
import pytensor.tensor as pt
import jax
from jax import lax, random
import jax.numpy as jnp
import numpyro
import numpyro.distributions as dist
import numpyro.handlers as handl
from numpyro.infer import MCMC, NUTS, SVI, Predictive, Trace_ELBO, autoguide
import optax
rng = np.random.default_rng(0)
numpyro.util.set_host_device_count(4)
rng_key = random.PRNGKey(0)
# GROUND TRUTH
w = 0.015
pareto_alpha = 0.5
pareto_m = 0.007
pareto_shift = 0.008
skewnorm_mu = -0.5
skewnorm_sigma = 0.8
skewnorm_alpha = 3.8
## GENERATE DATA
N = 10_000
_w = bernoulli(p=w).rvs(size=N, random_state=rng)
pareto_rvs = pareto(b=pareto_alpha, loc=pareto_shift, scale=pareto_m).rvs(size=N, random_state=rng)
logskewnorm_rvs = np.exp(skewnorm(a=skewnorm_alpha, loc=skewnorm_mu, scale=skewnorm_sigma).rvs(size=N, random_state=rng))
rvs = _w * pareto_rvs + (1 - _w) * logskewnorm_rvs
# PYMC MODEL
def _shiftedpareto_dist(alpha, m, shift, size):
return pm.Pareto.dist(alpha=alpha, m=m, size=size) + shift
def _logskewnorm_dist(mu, sigma, alpha, size):
return pt.exp(pm.SkewNormal.dist(mu=mu, sigma=sigma, alpha=alpha, size=size))
with pm.Model() as pymc_model:
latent_pareto_alpha = pm.TruncatedNormal("latent_pareto_alpha", mu=2, sigma=2, lower=1e-2, upper=6.0)
latent_pareto_m = pm.TruncatedNormal("latent_pareto_m", mu=0.0, sigma=0.1, lower=1e-3, upper=0.2)
latent_pareto_shift = pm.Deterministic("latent_pareto_shift", rvs.min() - latent_pareto_m)
leftoutlier_dist = pm.CustomDist.dist(
latent_pareto_alpha, latent_pareto_m, latent_pareto_shift,
dist=_shiftedpareto_dist,
class_name="ShiftedPareto")
latent_skewnorm_alpha = pm.TruncatedNormal("latent_skewnorm_alpha", mu=3, sigma=2, lower=-1, upper=7)
latent_skewnorm_mu = mu = pm.TruncatedNormal("latent_skewnorm_mu", mu=0.0, sigma=1, lower=-3, upper=3)
latent_skewnorm_sigma = pm.HalfNormal("latent_skewnorm_sigma", 1)
main_dist = pm.CustomDist.dist(
latent_skewnorm_mu, latent_skewnorm_sigma, latent_skewnorm_alpha,
dist=_logskewnorm_dist,
class_name="LogSkewNormal")
latent_w = pm.Dirichlet('latent_w', a=np.array([1, 30]))
obs = pm.Mixture(
'obs',
w=latent_w,
comp_dists=[leftoutlier_dist, main_dist],
observed=rvs)
# PYMC PRIOR PREDICTIVE
with pymc_model:
pymc_prior_pred = pm.sample_prior_predictive(500)
pymc_prior_obs = pymc_prior_pred.prior_predictive["obs"].values.reshape(-1)
# PYMC MCMC
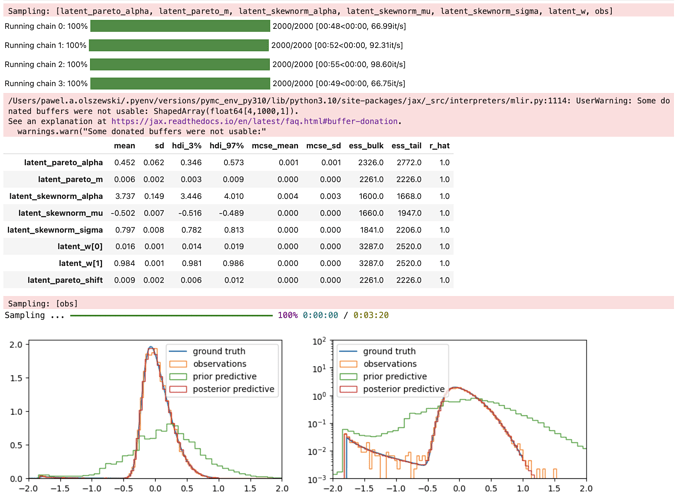
with pymc_model:
pymc_trace = pm.sample(nuts_sampler="numpyro")
display(pm.summary(pymc_trace))
# PYMC POSTERIOR PREDICTIVE
with pymc_model:
pymc_post_pred = pm.sample_posterior_predictive(pymc_trace)
pymc_post_obs = pymc_post_pred.posterior_predictive["obs"].values.reshape(-1)
# PLOT GROUND TRUTH, OBSERVATIONS, PYMC PRIOR- AND POSTERIOR PREDICTIVES,
# use log-scale(s)
x = np.linspace(0, 10, 3_000)[1:]
y = (
w * pareto(b=pareto_alpha, loc=pareto_shift, scale=pareto_m).pdf(x)
+ (1 - w) * skewnorm(a=skewnorm_alpha, loc=skewnorm_mu, scale=skewnorm_sigma).pdf(np.log(x))/x)
rvs_p99 = rvs[rvs < np.percentile(rvs, 99)]
pymc_prior_obs_p999 = pymc_prior_obs[(pymc_prior_obs < np.percentile(pymc_prior_obs, 99.9))]
pymc_post_obs_p9999 = pymc_post_obs[
(pymc_post_obs < np.percentile(pymc_post_obs, 99.99))]
fig, axs = plt.subplots(1, 2, figsize=(12, 3))
for ax in axs:
ax.plot(np.log10(x), y * x * np.log(10), label="ground truth");
ax.hist(np.log10(rvs), density=True, bins=100, histtype="step", label="observations");
ax.hist(np.log10(pymc_prior_obs_p999), density=True, bins=100, histtype="step", label="prior predictive");
ax.hist(np.log10(pymc_post_obs_p9999), density=True, bins=100, histtype="step", label="posterior predictive");
ax.legend();
ax.set_xlim(-2, 2);
axs[1].set_yscale("log");
axs[1].set_ylim(1e-3, 1e2);
# NUMPYRO MODEL
class ShiftedPareto(dist.TransformedDistribution):
arg_constraints = {"scale": dist.constraints.positive, "alpha": dist.constraints.positive}
reparametrized_params = ["scale", "alpha"]
def __init__(self, loc, scale, alpha, *, validate_args=None):
self.loc, self.scale, self.alpha = dist.util.promote_shapes(loc, scale, alpha)
batch_shape = lax.broadcast_shapes(jnp.shape(loc), jnp.shape(scale), jnp.shape(alpha))
loc, scale, alpha = (
jnp.broadcast_to(loc, batch_shape),
jnp.broadcast_to(scale, batch_shape),
jnp.broadcast_to(alpha, batch_shape),
)
base_dist = dist.Exponential(alpha)
transforms = [
dist.transforms.ExpTransform(),
dist.transforms.AffineTransform(loc=loc, scale=scale)
]
super(ShiftedPareto, self).__init__(base_dist, transforms, validate_args=validate_args)
@dist.constraints.dependent_property(is_discrete=False, event_dim=0)
def support(self):
return dist.constraints.greater_than(self.loc + self.scale)
class SkewNormal(dist.Distribution):
arg_constraints = {"mu": dist.constraints.real, "sigma": dist.constraints.positive, "alpha": dist.constraints.real}
support = dist.constraints.real
reparametrized_params = ["mu", "sigma", "alpha"]
def __init__(self, mu=0.0, sigma=1.0, alpha=0.0, *, validate_args=None):
self.mu, self.sigma, self.alpha = dist.util.promote_shapes(mu, sigma, alpha)
self.tau = 1 / self.sigma ** 2
batch_shape = lax.broadcast_shapes(jnp.shape(mu), jnp.shape(sigma), jnp.shape(alpha))
super(SkewNormal, self).__init__(
batch_shape=batch_shape, validate_args=validate_args
)
@dist.util.validate_sample
def log_prob(self, value):
# https://github.com/pymc-devs/pymc/blob/36cca5b623dda4e75ac677c9d8ad6a321a2e72c2/pymc/distributions/continuous.py#L3149
return (
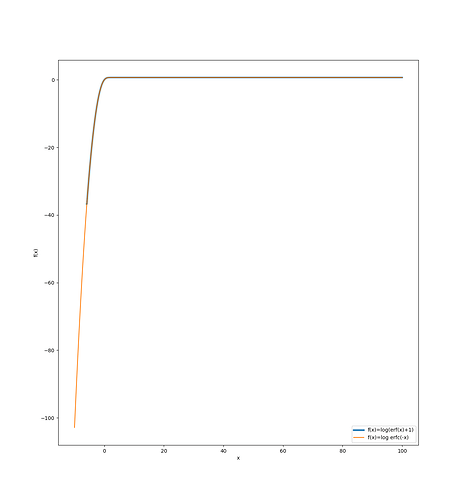
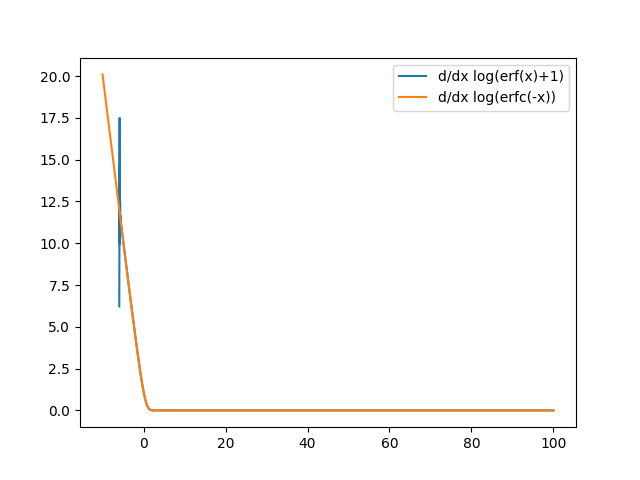
jnp.log1p(jax.scipy.special.erf((value - self.mu) / self.sigma * self.alpha / jnp.sqrt(2)))
+ (-self.tau * (value - self.mu) ** 2 + jnp.log(self.tau / jnp.pi / 2.0)) / 2.0
)
def sample(self, key, sample_shape=()):
# https://github.com/scipy/scipy/blob/92d2a8592782ee19a1161d0bf3fc2241ba78bb63/scipy/stats/_continuous_distns.py#L9422
assert numpyro.util.is_prng_key(key)
key_1, key_2 = random.split(key)
shape = sample_shape + self.batch_shape + self.event_shape
u0 = random.normal(key_1, shape=shape)
v = random.normal(key_2, shape=shape)
d = self.alpha/jnp.sqrt(1 + self.alpha**2)
u1 = d*u0 + v * jnp.sqrt(1 - d**2)
_rvs = jnp.where(u0 >= 0, u1, -u1)
rvs = _rvs * self.sigma + self.mu
return rvs
class LogSkewNormal(dist.TransformedDistribution):
arg_constraints = {"mu": dist.constraints.real, "sigma": dist.constraints.positive, "alpha": dist.constraints.real}
support = dist.constraints.positive
reparametrized_params = ["mu", "sigma", "alpha"]
def __init__(self, mu=0.0, sigma=1.0, alpha=0.0, *, validate_args=None):
base_dist = SkewNormal(mu, sigma, alpha)
self.mu, self.sigma, self.alpha = base_dist.mu, base_dist.sigma, base_dist.alpha
super(LogSkewNormal, self).__init__(
base_dist, dist.transforms.ExpTransform(), validate_args=validate_args
)
def numpyro_model(n_obs):
latent_pareto_alpha = numpyro.sample("latent_pareto_alpha", dist.TruncatedNormal(loc=2, scale=2, low=1e-2, high=6))
latent_pareto_m = numpyro.sample("latent_pareto_m", dist.TruncatedNormal(loc=0, scale=0.1, low=1e-3, high=0.2))
latent_pareto_shift = numpyro.deterministic("latent_pareto_shift", rvs.min() - latent_pareto_m)
leftoutlier_dist = ShiftedPareto(
loc=latent_pareto_shift, scale=latent_pareto_m, alpha=latent_pareto_alpha)
latent_skewnorm_alpha = numpyro.sample("latent_skewnorm_alpha", dist.TruncatedNormal(loc=3, scale=2, low=-1, high=7))
latent_skewnorm_mu = numpyro.sample("latent_skewnorm_mu", dist.TruncatedNormal(loc=0, scale=1, low=-3, high=3))
latent_skewnorm_sigma = numpyro.sample("latent_skewnorm_sigma", dist.HalfNormal(scale=1))
main_dist = LogSkewNormal(
mu=latent_skewnorm_mu, sigma=latent_skewnorm_sigma, alpha=latent_skewnorm_alpha)
latent_w = numpyro.sample("latent_w", dist.Dirichlet(concentration=jnp.array([1.0, 30.0])))
with numpyro.plate("obs_plate", n_obs):
obs = numpyro.sample(
"obs",
dist.MixtureGeneral(
mixing_distribution=dist.Categorical(probs=latent_w),
component_distributions=[leftoutlier_dist, main_dist],
support=dist.constraints.positive))
# NUMPYRO PRIOR PREDICTIVE
rng_key, rng_key_ = random.split(rng_key)
numpyro_prior_pred = Predictive(numpyro_model, num_samples=10_000)(rng_key_, n_obs=1)
numpyro_prior_obs = numpyro_prior_pred["obs"].reshape(-1)
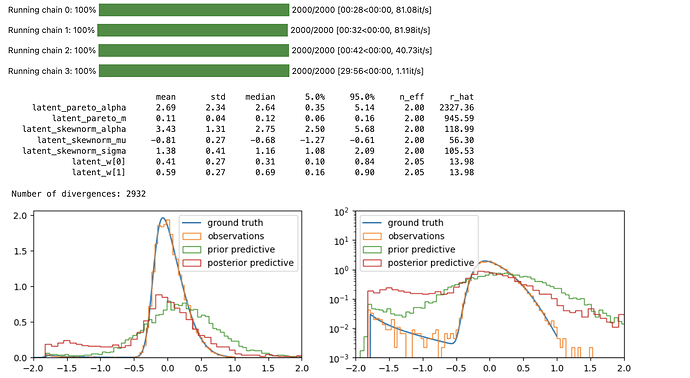
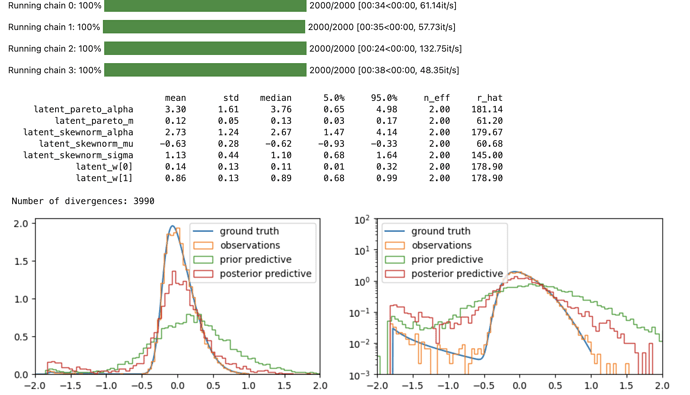
# NUMPYRO MCMC
numpyro_mcmc = MCMC(
sampler=NUTS(handl.condition(numpyro_model, {"obs": rvs})),
num_warmup=1_000,
num_samples=1_000,
num_chains=4,
)
rng_key, rng_key_ = random.split(rng_key)
numpyro_mcmc.run(rng_key_, n_obs=len(rvs))
numpyro_mcmc.print_summary()
numpyro_mcmc_samples = numpyro_mcmc.get_samples(group_by_chain=True)
# NUMPYRO POSTERIOR PREDICTIVE
rng_key, rng_key_ = random.split(rng_key)
numpyro_post_pred = Predictive(numpyro_model, posterior_samples=numpyro_mcmc_samples)(rng_key_, n_obs=1)
numpyro_post_obs = numpyro_post_pred["obs"].reshape(-1)
# PLOT GROUND TRUTH, OBSERVATIONS, NUMPYRO PRIOR- AND POSTERIOR PREDICTIVES,
# use log-scale(s)
rvs_p99 = rvs[rvs < np.percentile(rvs, 99)]
numpyro_prior_obs_p999 = numpyro_prior_obs[(numpyro_prior_obs < np.percentile(numpyro_prior_obs, 99.9))]
numpyro_post_obs_p9999 = numpyro_post_obs[
(numpyro_post_obs < np.percentile(numpyro_post_obs, 99.99))]
fig, axs = plt.subplots(1, 2, figsize=(12, 3))
for ax in axs:
ax.plot(np.log10(x), y * x * np.log(10), label="ground truth");
ax.hist(np.log10(rvs), density=True, bins=100, histtype="step", label="observations");
ax.hist(np.log10(numpyro_prior_obs_p999), density=True, bins=100, histtype="step", label="prior predictive");
ax.hist(np.log10(numpyro_post_obs_p9999), density=True, bins=100, histtype="step", label="posterior predictive");
ax.legend();
ax.set_xlim(-2, 2);
axs[1].set_yscale("log");
axs[1].set_ylim(1e-3, 1e2);