Hi all,
I am working on a toy example that learns the influence of binary configuration options on some non-functional property with the influences being my random variables. Although I implemented my artificial toy target function such that the influence of option A varies largely, NUTS infers all influences with the same variance.
This is my code:
def ask_oracle(config):
base = 20
a, b, c = config
influence_a = scipy.stats.norm(5, 3).rvs(1)[0]
influence_b = 0.5
influence_c = 2
nfp = influence_a * a + influence_b * b + influence_c * c + base
return nfp
Here, we define that influence_a should be normal-distributed with mean 5 and stddev 3.
This is my model:
def model(a, b, c, obs=None):
mean = 0
var = 15
influence_a = numpyro.sample("influence_a", npdist.Normal(mean, var))
influence_b = numpyro.sample("influence_b", npdist.Normal(mean, var))
influence_c = numpyro.sample("influence_c", npdist.Normal(mean, var))
base = numpyro.sample("base", npdist.HalfNormal(30))
result = base + a * influence_a + b * influence_b + c * influence_c
error_var = numpyro.sample("error", npdist.HalfNormal(1.0))
with numpyro.plate("data", len(a)):
obs = numpyro.sample("nfp", npdist.Normal(result, error_var), obs=obs)
return obs
And this is my experiment:
def main():
configs = list(itertools.product([True, False], repeat=3))
configs = configs * 20 # simulating repeated measurements
python_random.shuffle(configs)
nfp = jnp.atleast_1d(list(map(ask_oracle, configs)))
X = jnp.atleast_2d(configs)
X = MinMaxScaler().fit_transform(X)
nuts_kernel = npNUTS(model)
n_chains = 3
mcmc = npMCMC(nuts_kernel, num_samples=2000,
num_warmup=5000, progress_bar=False, num_chains=n_chains)
rng_key = random.PRNGKey(0)
mcmc.run(rng_key, X[:, 0], X[:, 1], X[:, 2], obs=nfp)
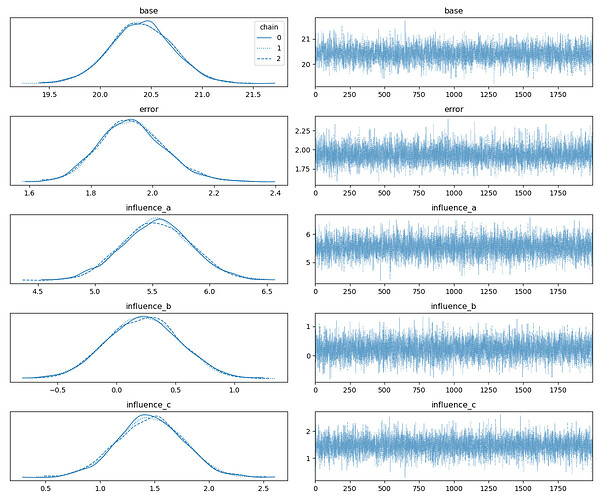
mcmc.print_summary()
az_data = az.from_numpyro(mcmc, num_chains=n_chains)
az.plot_trace(az_data, legend=True,)
This is the output:
mean std median 5.0% 95.0% n_eff r_hat
base 20.41 0.31 20.41 19.93 20.93 3693.94 1.00
error 1.93 0.11 1.93 1.75 2.10 5030.02 1.00
influence_a 5.54 0.30 5.54 5.02 6.02 4867.74 1.00
influence_b 0.23 0.31 0.23 -0.26 0.76 4564.49 1.00
influence_c 1.46 0.30 1.46 0.98 1.97 4685.77 1.00
I assumed that MCMC would assign more uncertainty to option A’s influence instead of the other RVs in order to approach the posterior more closely.
What can I do to detect this varying uncertainty? Is this generally possible?