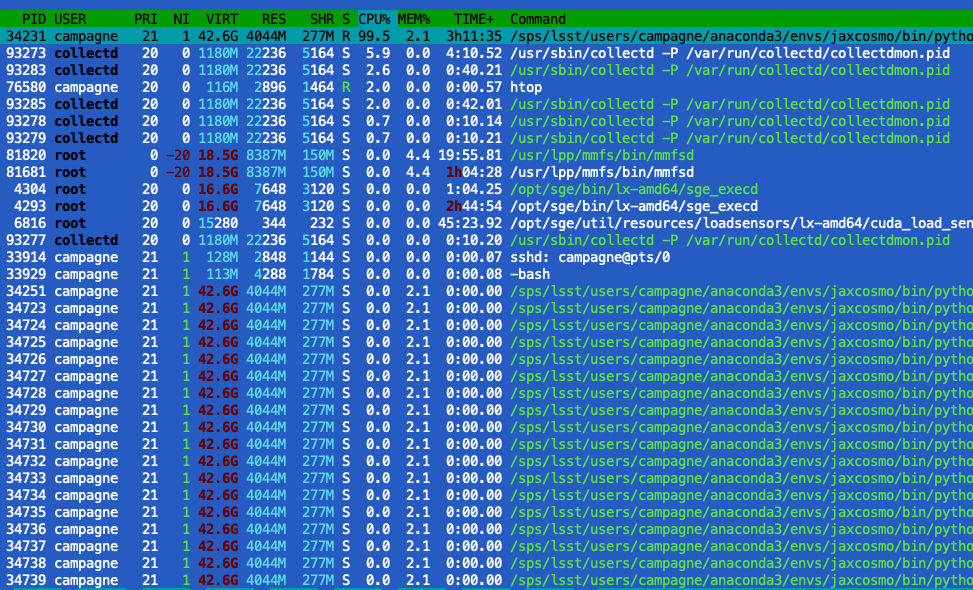
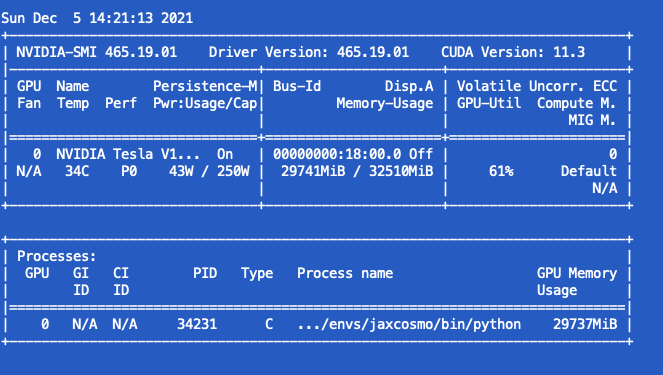
Hello,
I am running a CPU (Linux) + GPU-V100 hardware to perform SVI+NeuraTransform->MCMC-NUTS
Well, what is puzziling me is that
- the SVI uses a lot of threads (~40) which non of them uses the CPU while a thread is using 100% of the CPU and little GPU
- For the Neutra it is the same in terms of CPU & thread, while the GPU is used a 60% or so.
I wander if there are something that I have missed to make the CPU threads working and the GPU more efficiently used.
Here are some infos on my packages and some snippets
_libgcc_mutex 0.1 conda_forge conda-forge
_openmp_mutex 4.5 1_llvm conda-forge
absl-py 1.0.0 pyhd8ed1ab_0 conda-forge
astropy 4.3.1 py38h09021b7_0
brotli 1.0.9 he6710b0_2
ca-certificates 2021.10.26 h06a4308_2
certifi 2021.10.8 py38h06a4308_0
colorama 0.4.4 pyh9f0ad1d_0 conda-forge
cudatoolkit 11.3.1 h2bc3f7f_2
cudnn 8.2.1 cuda11.3_0
cycler 0.10.0 py38_0
dbus 1.13.18 hb2f20db_0
expat 2.4.1 h2531618_2
fontconfig 2.13.1 h6c09931_0
fonttools 4.25.0 pyhd3eb1b0_0
freetype 2.11.0 h70c0345_0
giflib 5.2.1 h7b6447c_0
glib 2.69.1 h5202010_0
gst-plugins-base 1.14.0 h8213a91_2
gstreamer 1.14.0 h28cd5cc_2
icu 58.2 he6710b0_3
jax 0.2.25 pyhd8ed1ab_0 conda-forge
jaxlib 0.1.73+cuda11.cudnn82 pypi_0 pypi
jpeg 9d h7f8727e_0
kiwisolver 1.3.1 py38h2531618_0
lcms2 2.12 h3be6417_0
ld_impl_linux-64 2.35.1 h7274673_9
libblas 3.9.0 12_linux64_openblas conda-forge
libcblas 3.9.0 12_linux64_openblas conda-forge
libffi 3.3 he6710b0_2
libgcc-ng 11.2.0 h1d223b6_11 conda-forge
libgfortran-ng 11.2.0 h69a702a_11 conda-forge
libgfortran5 11.2.0 h5c6108e_11 conda-forge
liblapack 3.9.0 12_linux64_openblas conda-forge
libopenblas 0.3.18 pthreads_h8fe5266_0 conda-forge
libpng 1.6.37 hbc83047_0
libstdcxx-ng 11.2.0 he4da1e4_11 conda-forge
libtiff 4.2.0 h85742a9_0
libuuid 1.0.3 h7f8727e_2
libwebp 1.2.0 h89dd481_0
libwebp-base 1.2.0 h27cfd23_0
libxcb 1.14 h7b6447c_0
libxml2 2.9.12 h03d6c58_0
llvm-openmp 12.0.1 h4bd325d_1 conda-forge
lz4-c 1.9.3 h295c915_1
matplotlib 3.4.3 py38h06a4308_0
matplotlib-base 3.4.3 py38hbbc1b5f_0
munkres 1.1.4 py_0
ncurses 6.3 h7f8727e_2
numpy 1.21.4 py38he2449b9_0 conda-forge
numpyro 0.8.0 pyhd8ed1ab_0 conda-forge
olefile 0.46 pyhd3eb1b0_0
openssl 1.1.1l h7f8727e_0
opt_einsum 3.3.0 pyhd8ed1ab_1 conda-forge
pcre 8.45 h295c915_0
pillow 8.4.0 py38h5aabda8_0
pip 21.2.4 py38h06a4308_0
pyerfa 2.0.0 py38h27cfd23_0
pyparsing 3.0.4 pyhd3eb1b0_0
pyqt 5.9.2 py38h05f1152_4
python 3.8.12 h12debd9_0
python-dateutil 2.8.2 pyhd3eb1b0_0
python-flatbuffers 2.0 pyhd8ed1ab_0 conda-forge
python_abi 3.8 2_cp38 conda-forge
pyyaml 6.0 py38h7f8727e_1
qt 5.9.7 h5867ecd_1
readline 8.1 h27cfd23_0
scipy 1.7.3 py38h56a6a73_0 conda-forge
setuptools 58.0.4 py38h06a4308_0
sip 4.19.13 py38he6710b0_0
six 1.16.0 pyh6c4a22f_0 conda-forge
sqlite 3.36.0 hc218d9a_0
tk 8.6.11 h1ccaba5_0
tornado 6.1 py38h27cfd23_0
tqdm 4.62.3 pyhd8ed1ab_0 conda-forge
typing_extensions 4.0.0 pyha770c72_0 conda-forge
wheel 0.37.0 pyhd3eb1b0_1
xz 5.2.5 h7b6447c_0
yaml 0.2.5 h7b6447c_0
zlib 1.2.11 h7b6447c_3
zstd 1.4.9 haebb681_0
Now my Numpyro model looks schematically like
def model_spl(obs=None):
# define 16 priors
# compute y=f(all_prior_variables)
return numpyro.sample('y', dist.MultivariateNormal(y,
precision_matrix=P,
covariance_matrix=C),
obs=obs)
with P&C matrices of size 2500x2500 defined before and are constant in my modelling.
Now, I adjust an approximation of the posteriors via SVI using the following snippet (PHASE 1)
guide = autoguide.AutoMultivariateNormal(model_spl,
init_loc_fn=numpyro.infer.init_to_value(values=true_param))
optimizer = numpyro.optim.Adam(5e-3)
svi = SVI(model_spl, guide,optimizer,loss=Trace_ELBO())
from functools import partial
def body_fn(i,carry):
svi_state, svi_state_best, losses = carry
svi_state, loss =svi.update(svi_state,cl_obs)
def update_fn(x):
return losses.at[i].set(loss), svi_state
def keep_fn(x):
return losses.at[i].set(losses[i-1]), svi_state_best
losses, svi_state_best = jax.lax.cond(loss<losses[i-1],update_fn,keep_fn,None)
return (svi_state, svi_state_best, losses)
svi_state = svi.init(jax.random.PRNGKey(42),cl_obs)
num_steps=5_000
losses = np.zeros(num_steps)
losses = losses.at[0].set(1e10)
svi_state_best = svi_state
carry = (svi_state,svi_state_best,losses)
carry = jax.lax.fori_loop(1,num_steps,body_fn,carry)
print("Save loss")
np.save('loss_shear_nc_DEBUG.npy',carry[2]) #step decrease of the loss
Now, using NeuraParam I perform a MCMC/NUTS using
from numpyro.infer.reparam import NeuTraReparam
from numpyro.infer import MCMC, NUTS, init_to_sample
neutra = NeuTraReparam(guide, svi.get_params(carry[1]))
neutra_model = neutra.reparam(model_spl)
nuts_kernel = NUTS(neutra_model,
init_strategy=numpyro.infer.init_to_median(),
dense_mass=True,
max_tree_depth=5)
mcmc_neutra = MCMC(nuts_kernel, num_warmup=1_000,
num_samples=8_000,
num_chains=1,progress_bar=False)
print('NUTS neutra')
mcmc_neutra.run(jax.random.PRNGKey(42), cl_obs)
mcmc_neutra.print_summary()
zs = mcmc_neutra.get_samples()["auto_shared_latent"]
samples_nuts_neutra = neutra.transform_sample(zs)
print("Save Neutra samples")
np.save('neutra_samples_shear_nc_dense_DEBUG.npy',samples_nuts_neutra)
Thanks for your attention.