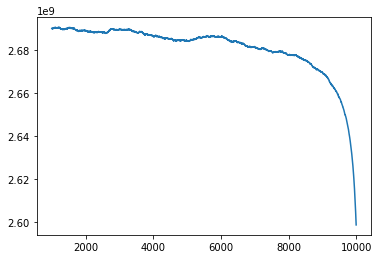
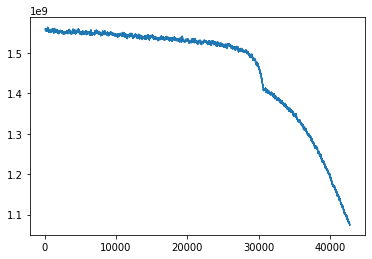
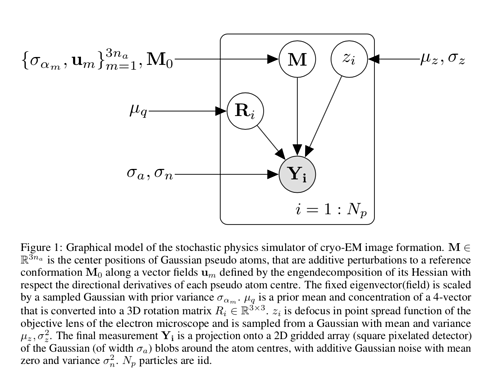
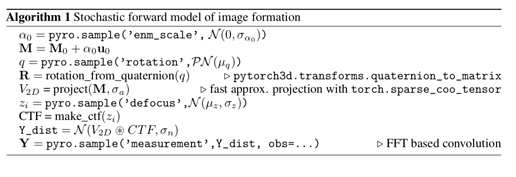
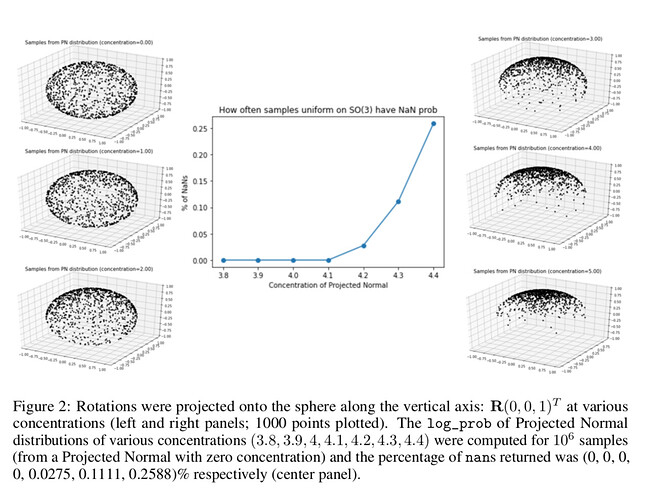
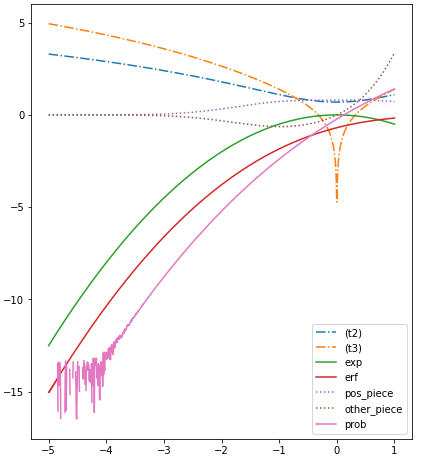
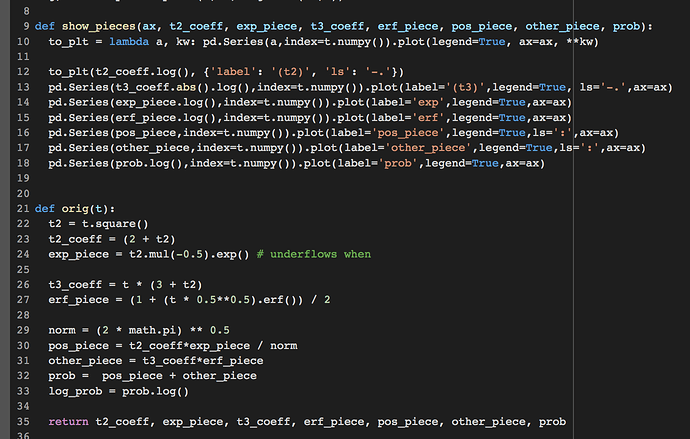
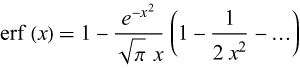This is the error
/usr/local/lib/python3.7/dist-packages/pyro/poutine/trace_struct.py:286: UserWarning: Encountered NaN: log_prob_sum at site 'quaternions'
site["log_prob_sum"], "log_prob_sum at site '{}'".format(name)
/usr/local/lib/python3.7/dist-packages/pyro/infer/trace_elbo.py:158: UserWarning: Encountered NaN: loss
warn_if_nan(loss, "loss")
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
/usr/local/lib/python3.7/dist-packages/pyro/poutine/trace_messenger.py in __call__(self, *args, **kwargs)
173 try:
--> 174 ret = self.fn(*args, **kwargs)
175 except (ValueError, RuntimeError) as e:
19 frames
ValueError: Expected parameter concentration (Tensor of shape (500, 2, 4)) of distribution ProjectedNormal(concentration: torch.Size([500, 2, 4])) to satisfy the constraint IndependentConstraint(Real(), 1), but found invalid values:
tensor([[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
...,
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]]], device='cuda:0', grad_fn=<MulBackward0>)
The above exception was the direct cause of the following exception:
ValueError Traceback (most recent call last)
<decorator-gen-53> in time(self, line, cell, local_ns)
<timed exec> in <module>()
/usr/local/lib/python3.7/dist-packages/torch/distributions/distribution.py in __init__(self, batch_shape, event_shape, validate_args)
54 if not valid.all():
55 raise ValueError(
---> 56 f"Expected parameter {param} "
57 f"({type(value).__name__} of shape {tuple(value.shape)}) "
58 f"of distribution {repr(self)} "
ValueError: Expected parameter concentration (Tensor of shape (500, 2, 4)) of distribution ProjectedNormal(concentration: torch.Size([500, 2, 4])) to satisfy the constraint IndependentConstraint(Real(), 1), but found invalid values:
tensor([[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
...,
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]],
[[nan, nan, nan, nan],
[nan, nan, nan, nan]]], device='cuda:0', grad_fn=<MulBackward0>)
Trace Shapes:
Param Sites:
net_rot$$$cnn_layers.0.weight 32 1 3 3
net_rot$$$cnn_layers.0.bias 32
net_rot$$$cnn_layers.2.weight 32 32 3 3
net_rot$$$cnn_layers.2.bias 32
net_rot$$$cnn_layers.5.weight 64 32 3 3
net_rot$$$cnn_layers.5.bias 64
net_rot$$$cnn_layers.7.weight 64 64 3 3
net_rot$$$cnn_layers.7.bias 64
net_rot$$$cnn_layers.10.weight 128 64 3 3
net_rot$$$cnn_layers.10.bias 128
net_rot$$$cnn_layers.12.weight 128 128 3 3
net_rot$$$cnn_layers.12.bias 128
net_rot$$$linear_layers.0.weight 512 2048
net_rot$$$linear_layers.0.bias 512
net_rot$$$linear_layers.2.weight 512 512
net_rot$$$linear_layers.2.bias 512
net_rot$$$linear_layers.4.weight 12 512
net_rot$$$linear_layers.4.bias 12
Sample Sites:
mini_batch dist |
value 500 |
dfs dist 500 |
value 500 |