Hello. everyone!
I’m new in pyro and bayesian programming, so I generated some data just like in model under iarange and tried to find parameters via SVI. Here data2model is a tensor with classes {0,…,4}, solutions is an array [10, 100, …]
def model(data2model):
loc = pyro.param('loc', 40 * torch.ones(data2model_len))
scale = pyro.param('scale', torch.ones(data2model_len), constraint=constraints.positive)
effort_coeff = pyro.param('effort_coeff', torch.tensor(1.), constraint=constraints.unit_interval)
with pyro.iarange('my_iarange', use_cuda=True):
a= pyro.sample('a', dist.Normal(loc, scale))
a= a.expand(
(len(solutions), data2model_len)).reshape((data2model_len, len(solutions)))
comfort = (a- torch.tensor(solutions).float().expand((data2model_len, -1))) *\
effort_coeff/abs(a)
softmax = torch.nn.Softmax(dim=0)
pyro.sample('picked', dist.Categorical(probs=softmax(comfort)), obs=data2model)
@config_enumerate(default="parallel")
def guide(data):
# loc = pyro.param('loc', 40 * torch.ones(data2model_len))
# scale = pyro.param('scale', torch.ones(data2model_len), constraint=constraints.positive)`
# with this and commented loc and scale below, sigma goes to 0. Now model converges to guide
# (loc and scale become 25 and 3)
with pyro.iarange('my_iarange', use_cuda=True):
loc = 25 * torch.ones(data2model_len)
scale = 3 * torch.ones(data2model_len)
a= pyro.sample('a', dist.Normal(loc, scale))
prior = torch.tensor([0.2, 0.1, 0.05, 0.05, 0.6]).expand((data2model_len, 5))
assignment_probs = pyro.param('assignment_probs', prior, constraint=constraints.unit_interval)
picked_prior = dist.Categorical(assignment_probs)
pyro.sample('picked', picked_prior, infer={'is_auxiliary': True})
optim = pyro.optim.Adam({'lr': 1e-1})
inference = SVI(model, guide, optim, loss=TraceEnum_ELBO(max_iarange_nesting=1))
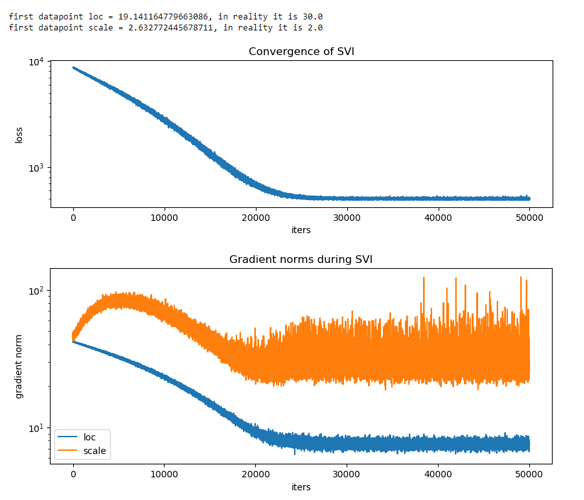
My generated data was sampled from normal(30, 2) and transformed. I have 2 different behaviors of my model (see comments in code). What can I do to successfully find real mu and sigma for my normal distribution? Also what does ‘is_auxiliary’ parameter does?
P.S. please connect login via fb or github on forum