Hi, I am experimenting with GMM for a case where there are a set of 3D points and only three unique coordinates. So, my data is
torch.tensor([
[0.2784, -0.3830, 0.8980],
[0.2784, -0.3830, 0.8980],
[0.2784, -0.3830, 0.8980],
[0.2784, -0.3830, 0.8980],
[0.2784, -0.3830, 0.8980],
[0.2581, 0.4620, -1.2788],
[0.2581, 0.4620, -1.2788],
[0.2581, 0.4620, -1.2788],
[0.2581, 0.4620, -1.2788],
[0.2581, 0.4620, -1.2788],
[1.0734, 0.4766, 0.5579],
[1.0734, 0.4766, 0.5579],
[1.0734, 0.4766, 0.5579],
[1.0734, 0.4766, 0.5579],
[1.0734, 0.4766, 0.5579]
])
and the unique points are
a = [0.2784, -0.3830, 0.8980]
b = [0.2581, 0.4620, -1.2788]
c = [1.0734, 0.4766, 0.5579]
Here is how all data points look like
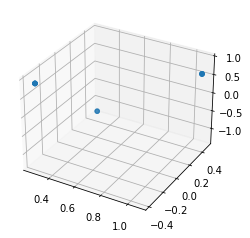
I expect the GMM to find three clusters whose centers are a, b, and c. I modified this tutorial for this experiment. My code is in https://gist.github.com/dilaragokay/e9e3d679e21db7711d2811f9e2904c03 (or here for seeing the cell outputs as well). As you can see in the notebook, all three inferred cluster centers are almost the same and do not reflect the data. I have tried many different hyperparameters, but the cluster centers are always away from the expected result. Is it possible that I am missing something in my approach?
Main differences with the tutorial:
- data is in 3D instead of 1D
- number of clusters is 3 instead of 2
- number of data points is 15 instead of 5
-
scaleis not sampled -
locsandobsare sampled fromMultivariateNormalinstead ofNormal - optimizer parameters
- initialization behavior of the autoguide