I’ve written a Bayesian model in numpyro and have been fitting the parameters with MCMC.
On the vast majority of chains (>95%) the posterior CIs contain the true parameters, the rhats are <1.05, and the chains seem to be sampling well. However, one in every 30 chains or so seems to collapse to a step size (~1e-7) significantly different from all other chains (~1e-3), with an acceptance rate (0.80) far lower than others (0.95).
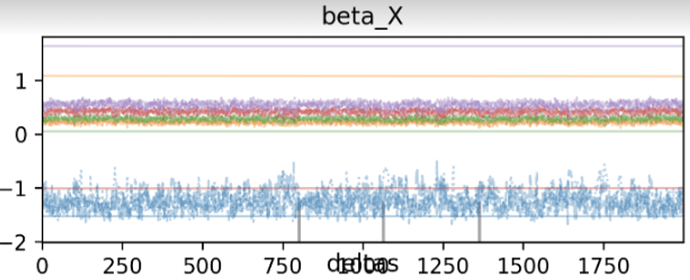
For instance, here’s what the MCMC chains look like in such a scenario:
sample: 100%|█████████████████████████████████████████████████████████████████| 2000/2000 [03:22<00:00, 9.90it/s, 255 steps of size 6.32e-03. acc. prob=0.95]
sample: 100%|█████████████████████████████████████████████████████████████████| 2000/2000 [03:30<00:00, 9.52it/s, 255 steps of size 7.97e-07. acc. prob=0.80]
sample: 100%|█████████████████████████████████████████████████████████████████| 2000/2000 [03:26<00:00, 9.70it/s, 255 steps of size 5.92e-03. acc. prob=0.96]
sample: 100%|█████████████████████████████████████████████████████████████████| 2000/2000 [03:24<00:00, 9.78it/s, 255 steps of size 8.89e-03. acc. prob=0.91]
mean std median 5.0% 95.0% n_eff r_hat
beta_X[0] -0.87 0.52 -1.08 -1.35 0.00 2.19 3.65
beta_X[1] 0.44 0.37 0.24 0.20 1.08 2.01 15.17
beta_X[2] 0.53 0.38 0.33 0.27 1.18 2.01 13.40
beta_X[3] 0.24 0.32 0.41 -0.30 0.48 2.03 8.85
beta_X[4] 0.31 0.42 0.53 -0.41 0.62 2.03 9.30
deltas[0] 0.21 0.16 0.13 0.10 0.49 2.01 13.04
deltas[1] 0.80 0.12 0.81 0.66 0.97 4.57 1.46
thresholds_in_p_space[0] 0.36 0.14 0.29 0.24 0.60 2.08 5.47
thresholds_in_p_space[1] 0.51 0.06 0.49 0.45 0.60 2.30 2.88
As you can tell, one chain is very different than the others and this is messing with the rhat value. Removing this chain brings the rhats back down to <1.05.
Looking at the trace plot, this outlier chain stays completely flat (see screenshot attached). I’m wondering if there’s any advice on how to deal with such issues. The posterior geometry shouldn’t be an issue here; I tried the same model with Stan and didn’t observe the same behavior.
I’m using 1000 warmups and 1000 samples. I’ve tried to play around with the max_tree_depth parameter. Decreasing it from 10 (default) to 8 seems to help, but the issue still doesn’t fully disappear.
I’m wondering if there’s a way to, e.g. lower bound the step size? Are there any other tips for such scenarios?