Following the sgt example I set my observed within the scan using handlers.condition, this works well for a standard distribution but fails for a transformed distribution. Is it possible with NUTS to set the sample for an observed, transformed distribution within a scan?
import numpy as np
import numpyro
import numpyro.distributions as dist
from numpyro.contrib.control_flow import scan
from numpyro.infer import NUTS, MCMC, init_to_median, Predictive
from numpyro.distributions.transforms import AffineTransform
from numpyro.infer.reparam import TransformReparam
from jax import random
import arviz as az
import jax.numpy as jnp
import jax
import pandas as pd
numpyro.set_host_device_count(4)
np.set_printoptions(suppress=True)
sample_params = {
'num_chains':4,
'num_warmup':1000,
'num_samples':1000,
'progress_bar':True,
}
#generate AR1 process
N, K = 200, 5
scale = 2.
z = np.random.RandomState(8).normal(scale=scale, size=(N, K))
theta = -0.2
y = np.zeros_like(z)
y[0, :] = z[0, :]
y[1:, :] = z[:-1, :]*theta + z[1:, :]
y = jnp.array(y, dtype=jnp.float32)
def model1(N, K, N_future, y):
scale = numpyro.sample('scale', dist.Exponential(1))
theta = numpyro.sample('theta', dist.Normal(0, 1))
def transition(y_prev, t):
loc = y_prev*theta
y_ = numpyro.sample('y', dist.Normal(loc=loc, scale=scale), )
return y_, y_
init_0 = y[0]
with numpyro.handlers.condition(data={'y': y}):
_, ys = scan(transition, init_0, jnp.arange(N + N_future), )
if N_future > 0:
numpyro.deterministic("y_forecast", ys[-N_future:])
hmc = MCMC(NUTS(model1, target_accept_prob=0.9, init_strategy=init_to_median), **sample_params)
hmc.run(random.PRNGKey(0), extra_fields=("z", "energy", "diverging"), N=N, K=K, y=y, N_future=0)
idata = az.from_numpyro(hmc, )
az.summary(idata)
def model2(N, K, N_future, y):
scale = numpyro.sample('scale', dist.Exponential(1))
theta = numpyro.sample('theta', dist.Normal(0, 1))
def transition(y_prev, t):
loc = y_prev*theta
y_ = numpyro.sample('y',
dist.TransformedDistribution(
dist.Normal(loc=loc, scale=1.),
AffineTransform(0, scale)
)
)
return y_, y_
init_0 = y[0]
with numpyro.handlers.condition(data={'y': y}):
_, ys = scan(transition, init_0, jnp.arange(N + N_future), )
if N_future > 0:
numpyro.deterministic("y_forecast", ys[-N_future:])
hmc = MCMC(NUTS(model2, target_accept_prob=0.9, init_strategy=init_to_median), **sample_params)
hmc.run(random.PRNGKey(0), extra_fields=("z", "energy", "diverging"), N=N, K=K, y=y, N_future=0)
idata = az.from_numpyro(hmc, )
az.summary(idata)
def model3(N, K, N_future, y):
scale = numpyro.sample('scale', dist.Exponential(1))
theta = numpyro.sample('theta', dist.Normal(0, 1))
def transition(y_prev, t):
loc = y_prev*theta
with numpyro.handlers.reparam(config={"y": TransformReparam()}):
y_ = numpyro.sample('y',
dist.TransformedDistribution(
dist.Normal(loc=loc, scale=1.),
AffineTransform(0, scale)
)
)
return y_, y_
init_0 = y[0]
with numpyro.handlers.condition(data={'y': y}):
_, ys = scan(transition, init_0, jnp.arange(N + N_future), )
if N_future > 0:
numpyro.deterministic("y_forecast", ys[-N_future:])
hmc = MCMC(NUTS(model3, target_accept_prob=0.9, init_strategy=init_to_median), **sample_params)
hmc.run(random.PRNGKey(0), extra_fields=("z", "energy", "diverging"), N=N, K=K, y=y, N_future=0)
idata = az.from_numpyro(hmc, )
az.summary(idata)
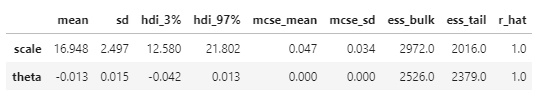
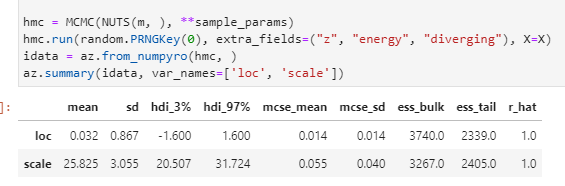
- model1() runs fine and and recovers the true parameters in my minimal working example.
- model2() fails with error:
NotImplementedError: Flatenning TransformedDistribution is only supported for some specific cases. Consider usingTransformReparamto convert this distribution to the base_dist, which is supported in most situtations. In addition, please reach out to us with your usage cases. - model3() runs without error but the condition handler doesn’t work to set the observed for
y
Here I am using AffineTransform which I could easily do without, as in model1(), but it’s just to display the error.