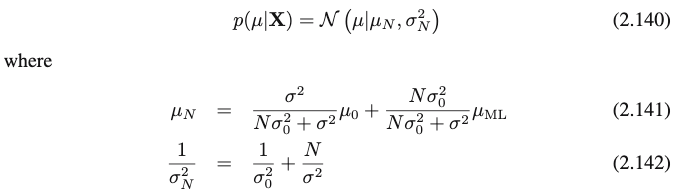Hello!
I’m toying with one of simplest examples involving Bayesian inference: modeling 1-dimensional data x using a Gaussian distribution N(x | μ, σ²) with prior over the mean N(μ | μ₀, σ₀²) and fixed variance
def model(x):
m0 = torch.tensor(0.0)
s0 = torch.tensor(10.0)
m = pyro.sample("m", dist.Normal(m0, s0))
s = torch.tensor(1.0)
pyro.sample("x", dist.Normal(m, s), obs=torch.tensor(x))
The posterior of the mean is set to a Gaussian distribution p(μ | x) ≅ q(μ) = N(μ | μ_q, σ²_q) by using the AutoNormal guide.
When training on a large synthetic dataset, the mean of the posterior μ_q does converge to the true mean of the data, but the variance σ²_q seems to stay constant – I was expecting it to decrease to 0 as more evidence is gathered.
...
900 | +3.068 | AutoNormal.locs.m: 2.98, AutoNormal.scales.m: 0.99
950 | +6.529 | AutoNormal.locs.m: 2.99, AutoNormal.scales.m: 1.04
1000 | +3.376 | AutoNormal.locs.m: 2.93, AutoNormal.scales.m: 1.09
1050 | +3.509 | AutoNormal.locs.m: 2.99, AutoNormal.scales.m: 1.08
1100 | +4.299 | AutoNormal.locs.m: 2.97, AutoNormal.scales.m: 1.13
...
Any ideas of what might be happening? I’m surely missing something fundamental, so any guidance is appreciated.
Here is the complete code:
import torch
import numpy as np
import pyro
import pyro.distributions as dist
from pyro.infer import SVI, Trace_ELBO
from pyro.infer.autoguide import guides
from pyro.optim import Adam
SEED = 1337
np.random.seed(SEED)
pyro.enable_validation(True)
def load_data(num_points, mu, sigma):
x = np.random.randn(num_points)
return mu + sigma * x
def model(x):
m0 = torch.tensor(0.0)
s0 = torch.tensor(10.0)
m = pyro.sample("m", dist.Normal(m0, s0))
s = torch.tensor(1.0)
pyro.sample("x", dist.Normal(m, s), obs=torch.tensor(x))
guide = guides.AutoNormal(model)
optimizer = Adam({"lr": 0.01})
svi = SVI(model, guide, optimizer, loss=Trace_ELBO())
data = load_data(5000, 3, 1)
fmt = lambda p: ", ".join(f"{k}: {v.item():6.2f}" for k, v in p.items())
for it, x in enumerate(data):
nll = svi.step(x)
if it % 50 == 0:
params = pyro.get_param_store()
print(f"{it:5d} | {nll:+8.3f} |", fmt(params))
params = pyro.get_param_store()
print("estimated: ", fmt(params))