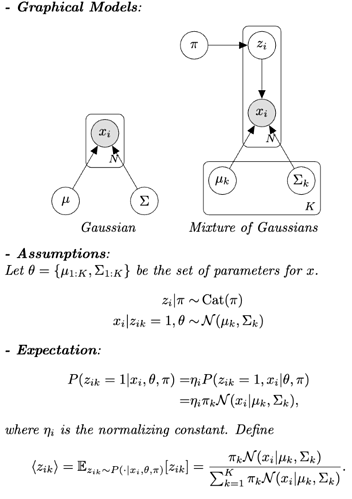
Hello everyone! I have read the GMM tutorial on the website, and I wanted to create a naive version of GMM where the mixture weights, means, covariance matrices are treated as learnable parameters instead of latent variables; in other words, no prior distributions are assumed for them. The model assumptions and the E-step are shown in the picture attached.
I think the major problem I had was I don’t know how to use the same set of parameters for model and guide. In pyro, it seems that the parameters (theta) for the generative process is defined in a model, while the parameters (phi) for the inference process is defined in a guide. In EM algorithm, we essentially use the same set of parameters for the E-step and M-step, and I don’t know how to reflect this in my code.
Here’s my model and guide:
def model(data, K=3):
num_obs,num_dim = data.shape
with pyro.plate("data", size=num_obs):
assignment = pyro.sample("assignment", dist.Categorical(torch.ones(K)/K))
means = torch.zeros(K, num_dim)
scale_tril = torch.eye(num_dim).repeat(K, 1, 1)
pyro.sample("obs", dist.MultivariateNormal(means[assignment], scale_tril=scale_tril[assignment]), obs=data)
def guide(data, K=3):
num_obs,num_dim = data.shape
# define learnable parameters: mixture weights, means, covariance matrices
weights = pyro.param("weights", torch.ones(K)/K, constraint=dist.constraints.simplex)
means = pyro.param("means", torch.randn(K, num_dim))
chol_factor = pyro.param("chol_factor", torch.eye(num_dim).repeat(K, 1, 1))
scale_diag = pyro.param("scale_diag", torch.ones(K, num_dim), constraint=dist.constraints.positive)
scale_tril = chol_factor @ torch.diag_embed(scale_diag)
with pyro.plate("data", size=num_obs):
# log(pi_k) + log(Normal(x_i | mu_k, Sigma_k))
log_probs = torch.stack([dist.MultivariateNormal(means[k], scale_tril=scale_tril[k]).log_prob(data) for k in range(K)], dim=-1) + torch.log(weights)
assignment_probs = torch.exp(log_probs - torch.logsumexp(log_probs, dim=1, keepdim=True))
assignment = pyro.sample("assignment", dist.Categorical(probs=assignment_probs))
Here is how I generate my data:
weights = np.array([2/8, 5/8, 1/8])
means = np.array([[1, 2],
[5, 6],
[3, 1]])
covariances = np.array([[[1.0, 0.2], [0.2, 1.5]],
[[3, -0.3], [-2, 9]],
[[1.2, 0.4], [0.4, 1.3]]])
def sample_from_mixture(weights, means, covariances, num_samples=100):
num_components = len(weights)
components = np.random.choice(num_components, size=num_samples, p=weights)
samples = np.array([
np.random.multivariate_normal(means[comp], covariances[comp])
for comp in components
])
return samples
num_samples = 1000
samples = sample_from_mixture(weights, means, covariances, num_samples)
samples = torch.tensor(samples, dtype=torch.float)
Here’s my training setup:
pyro.clear_param_store()
optim = Adam({"lr": 0.001, "betas": [0.8, 0.99]})
elbo = Trace_ELBO()
svi = SVI(model, guide, optim, elbo)
num_steps = 20000
for step in range(num_steps):
loss = svi.step(samples, K=3)
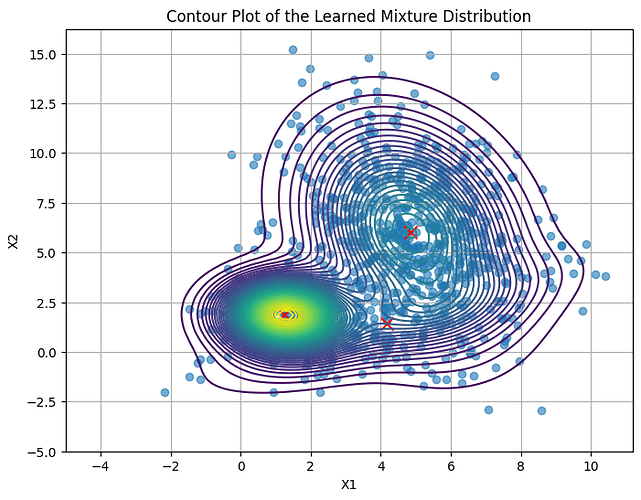
Training results: the training got stuck at a local minimum, and only one mode was learned. I think one of the reason is I defined prior distributions for z (“assignment”) and x (“obs”) in the model.