I am trying to fit a Bayesian model to a dataset of ~400K patient records with a multi-stage decision model. The model has a few key parameters: two decision thresholds (one threshold for each stage) and some regression coefficients.
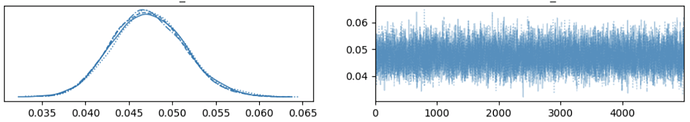
When I subsample the model to around 50-100k patient records, the model converges quite well. An example trace plot for thresholds[1] with 50k records is included below.
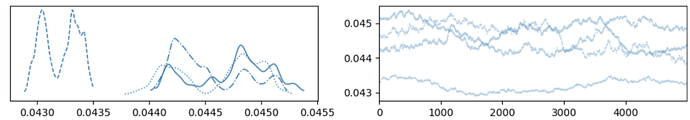
When I scale up to the full dataset, I no longer get convergence. The four chains don’t mix well at all. Here’s an example trace plot for thresholds[1] with 400k records:
I’m wondering if anyone has any pointers for what may be going wrong in this model, since all my parameters are global (so I don’t see why increasing the amount of data would cause the chains to mix poorly). I’ve included the model code below. I’ve tried different amounts of warmup (500, 1000, 5000) and samples (same numbers) but still observe the same discrepancy.
Any general intuitions for why such a problem may occur would be well appreciated!
def multi_stage_model(M, N, K, last_feature_idx_to_use, y, X, stage, beta_X=None):
# Priors for parameters
beta_X = numpyro.sample('beta_X', dist.Normal(0, 1).expand([N]))
logit_threshold_0 = numpyro.sample('logit_threshold_0', dist.Normal(-1, 1))
gap_0_1 = numpyro.sample('gap_0_1', dist.HalfNormal(jnp.sqrt(0.5)))
logit_thresholds_in_p_space = jnp.array([logit_threshold_0, logit_threshold_0 + gap_0_1])
thresholds_in_p_space = jax.scipy.special.expit(logit_thresholds_in_p_space)
threshold_0 = numpyro.deterministic('threshold_0', thresholds_in_p_space[0])
threshold_1 = numpyro.deterministic('threshold_1', thresholds_in_p_space[1])
deltas_0 = numpyro.sample('delta_0', dist.HalfNormal(jnp.sqrt(0.1)))
gap_delta = numpyro.sample('gap_delta', dist.HalfNormal(jnp.sqrt(0.3)))
deltas = jnp.array([deltas_0, deltas_0 + gap_delta])
delta_0 = numpyro.deterministic('delta_0_calculated', deltas[0])
delta_1 = numpyro.deterministic('delta_1_calculated', deltas[1])
# Loop over stages
for k in range(K):
# Compute phi for this stage using only the relevant features
phi = jax.scipy.special.expit(jnp.dot(X[:, :last_feature_idx_to_use[k]], beta_X[:last_feature_idx_to_use[k]]))
phi = jnp.clip(phi, 1e-6, 1-1e-6)
thresholds_in_p_space = jnp.clip(thresholds_in_p_space, 1e-6, 1-1e-6)
# Convert threshold from p-space to signal space for the current stage
log_ratio = jnp.log((phi / (1 - phi)) * ((1 - thresholds_in_p_space[k]) / thresholds_in_p_space[k]))
log_ratio = jnp.clip(log_ratio, -100, 100)
threshold_in_signal_space = (deltas[k]**2 - 2 * log_ratio) / (2 * deltas[k])
threshold_in_signal_space = jnp.clip(threshold_in_signal_space, -100, 100)
# Calculate probability mass above the threshold for the current stage
mass_above_threshold_pos = phi * (1 - jax.scipy.stats.norm.cdf(threshold_in_signal_space, loc=deltas[k], scale=1))
mass_above_threshold_neg = (1 - phi) * (1 - jax.scipy.stats.norm.cdf(threshold_in_signal_space, loc=0, scale=1))
mass_above_threshold_pos = jnp.clip(mass_above_threshold_pos, 1e-6, 1-1e-6)
mass_above_threshold_neg = jnp.clip(mass_above_threshold_neg, 1e-6, 1-1e-6)
search_rate = mass_above_threshold_pos + mass_above_threshold_neg
search_rate = jnp.clip(search_rate, 1e-6, 1-1e-6)
# Masking logic for the current stage
if k == 0:
stage_1_mask = (stage > 1) # Stage 1, only those who move on to stage 2 or beyond
numpyro.factor('stage_1_passing', jnp.sum(stage_1_mask * jnp.log(search_rate)))
numpyro.factor('stage_1_not_passing', jnp.sum(~stage_1_mask * jnp.log(1 - search_rate)))
else:
stage_passing_mask = (stage >= (k + 2)) # Those who moved on to the next stage
stage_not_passing_mask = (stage == k + 1) # Those who dropped out at this stage
# Calculate contributions only for those who passed the previous stages
numpyro.factor(f'stage_{k+1}_passing', jnp.sum(stage_passing_mask * jnp.log(search_rate)))
numpyro.factor(f'stage_{k+1}_not_passing', jnp.sum(stage_not_passing_mask * jnp.log(1 - search_rate)))
semi_final_stage_mask = (stage == K)
mass_below_threshold_neg = (1 - phi) * jax.scipy.stats.norm.cdf(threshold_in_signal_space, loc=0, scale=1)
mass_below_threshold_pos = phi * jax.scipy.stats.norm.cdf(threshold_in_signal_space, loc=deltas[K-1], scale=1)
search_rate_penultimate = mass_below_threshold_pos + mass_below_threshold_neg
search_rate_penultimate = jnp.clip(search_rate_penultimate, 1e-6, 1-1e-6)
hit_rate = mass_below_threshold_pos / search_rate_penultimate
hit_rate = jnp.clip(hit_rate, 1e-6, 1-1e-6)
with numpyro.handlers.mask(mask=semi_final_stage_mask):
numpyro.sample('obs_semi_final', dist.Bernoulli(probs=hit_rate), obs=y)
# Final outcome (y) for those who passed all stages
final_stage_mask = (stage == K + 1)
hit_rate = mass_above_threshold_pos / search_rate
hit_rate = jnp.clip(hit_rate, 1e-6, 1-1e-6)
# Carefully using the mask for final outcome observations
with numpyro.handlers.mask(mask=final_stage_mask):
numpyro.sample('obs', dist.Bernoulli(probs=hit_rate), obs=y)