Hello All.
I wanted to try variational Inference of Gaussian Mixture Model using pyro, I made the code referring following site.
Gaussian Mixture Model — Pyro Tutorials 1.8.6 documentation
I want to trace “Gradinet norm” of posterior parameter depending on iteration, I ran the following code.
for name, value in pyro.get_param_store().named_parameters():
value.register_hook(
lambda g, name=name: gradient_norms[name].append(g.norm().item())
)
But this code didn’t run well. I investigated this code and I found that “pyro.get_param_store().named_parameters()” has no parameters. Because I ran only “pyro.get_param_store().named_parameters()”,then the out put was “dict() ”
why “pyro.get_param_store().named_parameters()” didn’t include parameters?
[Execution environment], [Imported library], [Train data],[ model],[ guide], 【the code of just before “for name, value in pyro.get_param_store().named_parameters()” 】is following. If there is lack of the information, it is helpful for me to inform.
[Execution environment]
google colaboratory
[Imported library]
pip install pyro-ppl
import os
from collections import defaultdict
import torch
import numpy as np
import scipy.stats
from torch.distributions import constraints
from matplotlib import pyplot
%matplotlib inline
import pyro
import pyro.distributions as dist
from pyro import poutine
from pyro.infer.autoguide import AutoDelta
from pyro.optim import Adam
from pyro.infer import SVI, TraceEnum_ELBO, config_enumerate, infer_discrete
from tqdm import tqdm
smoke_test = "CI" in os.environ
assert pyro.__version__.startswith('1.8.6')
import matplotlib.animation as animation
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inlin
[Train data]
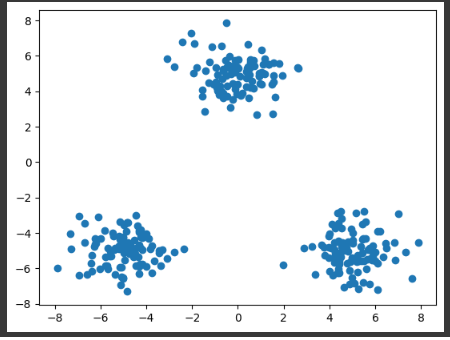
x1 = np.random.normal(size=(100, 2))
x1 += np.array([-5, -5])
x2 = np.random.normal(size=(100, 2))
x2 += np.array([5, -5])
x3 = np.random.normal(size=(100, 2))
x3 += np.array([0, 5])
x_train = np.vstack((x1, x2, x3))
x0, x1 = np.meshgrid(np.linspace(-10, 10, 100), np.linspace(-10, 10, 100))
x = np.array([x0, x1]).reshape(2, -1).T
plt.scatter(x_train[:, 0], x_train[:, 1])
x_train_ten = torch.from_numpy(x_train).clone()
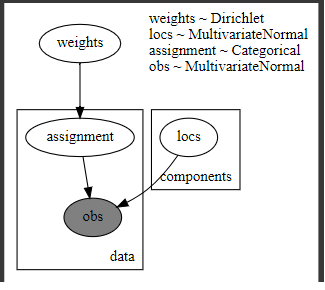
[model]
@config_enumerate
def gmm_pyro1(data,K=3):
# Global variables.
weights = pyro.sample("weights", dist.Dirichlet(0.5 * torch.ones(K)))
scales = torch.eye(2)
with pyro.plate("components", K):
locs = pyro.sample("locs", dist.MultivariateNormal(torch.zeros(2), scales))
with pyro.plate("data", len(data)):
# Local variables.
assignment = pyro.sample("assignment", dist.Categorical(weights))
pyro.sample("obs",dist.MultivariateNormal(locs[assignment], scales), obs=data)
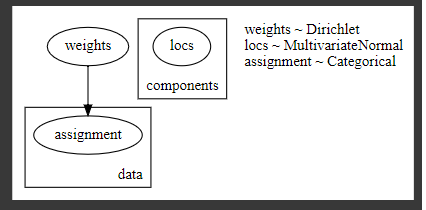
[guide]
Because I want the posterior distribution of parameter (“loc”,”weights”),I didn’t use AutoDelta and made custom guide.
@config_enumerate
def gmm_pyro1_guide(data,K=3):
# Global variables.
weights = pyro.sample("weights", dist.Dirichlet(0.5 * torch.ones(K)))
scales = torch.eye(2)
with pyro.plate("components", K):
#scale=pyro.sample("scale", dist.Wishart(torch.eye(2), torch.Tensor([2]))
locs = pyro.sample("locs", dist.MultivariateNormal(torch.zeros(2), scales))
with pyro.plate("data", len(data)):
# Local variables.
assignment = pyro.sample("assignment", dist.Categorical(weights))
【the code of just before “for name, value in pyro.get_param_store().named_parameters()” 】
pyro.clear_param_store()
gmm_pyro1_guide_pou=poutine.block(gmm_pyro1_guide, expose=["weights", "locs"])
optim = pyro.optim.Adam({"lr": 0.1, "betas": [0.8, 0.99]})
elbo = TraceEnum_ELBO(max_plate_nesting=1)
svi = SVI(gmm_pyro1,gmm_pyro1_guide_pou,optim,elbo)
svi.loss(gmm_pyro1,gmm_pyro1_guide_pou,x_train_ten)
gradient_norms = defaultdict(list)
for name, value in pyro.get_param_store().named_parameters():
print("name is:",name)
value.register_hook(
lambda g, name=name: gradient_norms[name].append(g.norm().item())
)