Hi,
I’m am trying to follow the neural hmm example to build a DBN for some predictive task (model description bellow).
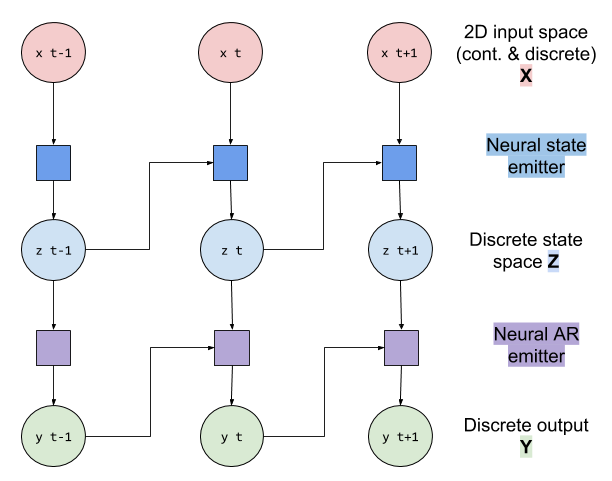
I am working with mostly discrete data, and using two MLP nn.Module modules (StateEmitter and Emitter) to model the states transition and the emission of the state space model.
The problem occurs after the first batch in the inference.
Code
The MLP modules look like
class Emitter(nn.Module):
...
def forward(self, y, z):
# Check dimension of y so this can be used with and without enumeration.
if y.dim() < 2:
y = y.unsqueeze(0)
# move to onehot representation
z_onehot = self.int2onehot(z, self.num_states, y.dtype, y.device).type(torch.float)
y_onehot = self.int2onehot(y, self.num_categories, y.dtype, y.device, add_batch_dim=True).type(torch.float)
# compute the linear projection of the onehot y_{t-1}. The onehot state vector z will be enumerated
# onehot vectors dim [batch_size, channels, length]
y_conv = self.relu(self.conv_y(y_onehot)).reshape(y.shape[:-1] + (-1,))
# add computed layer, project to y's (output) dimension and turn into probabilities
proposed_alpha = self.lin_hidden_to_y(self.lin_y_to_y_hidden(y_conv) + self.lin_z_to_z_hidden(z_onehot))
alpha = self.softmax(proposed_alpha)
return alpha
class GatedStateTransition(nn.Module):
...
def forward(self, w, z):
if w.dim() < 2:
w = w.unsqueeze(0)
# compute the gating function
_gate = self.relu(self.lin_gate_w_to_hidden(w))
gate = self.sigmoid(self.lin_gate_hidden_to_z(_gate))
_proposed_alpha = self.relu(self.lin_proposed_concentration_w_to_hidden(w))
proposed_alpha = self.lin_proposed_concentration_hidden_to_z(_proposed_alpha)
z_long = torch.Tensor([z]).type(torch.LongTensor) if not torch.is_tensor(z) else z
z_onehot = (
torch.zeros(z_long.shape[:-1] + (self.num_states,), dtype=w.dtype, device=w.device).scatter_(-1, z_long, 1))
alpha = self.softmax((1 - gate) * self.lin_z_to_concentration(z_onehot) + gate * proposed_alpha)
return alpha
def model(self, sequences, include_prior=True):
...
output_dim = output_seq[0].shape[1]
pyro.module("state_emitter", self.state_emitter)
pyro.module("ar_emitter", self.ar_emitter)
with poutine.mask(mask=include_prior):
probs_lat = pyro.sample("probs_lat",
dist.Dirichlet(
0.5 * torch.eye(self.num_states) + 0.5 / (self.num_states - 1)).to_event(1))
obs_plate = pyro.plate("obs", output_dim, dim=-1)
with pyro.plate("sequence_list", self.num_seqs, self.batch_size, dim=-2) as batch:
lengths = self.lengths[batch]
z = 0
y = torch.zeros(self.args.batch_size,1)
for t in pyro.markov(range(0, self.max_lenght if self.args.jit else self.lengths.max())):
with poutine.mask(mask=(t < lengths).unsqueeze(-1)):
emitted_x = pyro.sample(f"emit_x_{t}", dist.Categorical(self.state_emitter(input_seq[batch, t], z)[:,None,:]),
infer={"enumerate": "parallel"})
z = pyro.sample(f"z_{t}", dist.Categorical(probs_lat[emitted_x]),
infer={"enumerate": "parallel"})
with obs_plate:
y = pyro.sample(f"y_{t}", dist.Categorical(self.ar_emitter(y, z)).to_event(1), obs=output_seq[batch, t])
I am using TraceEnum_ELBO and AutoDelta(poutine.block(self._model, expose=["probs_lat"])) guide in SVI(model, guide, optim, elbo)
Questions
- The first batch goes fine, the second batch alters
emitted_xsize, where a dimension is added after each batch. The code fails in the second batch (in the last line of the modely=...) with the error
ValueError: Shape mismatch inside plate('sequence_list') at site y_0 dim -2, 30 vs 6. I can’t figure out why, but to blame a misuse of enumeration. - Do I use enumeration right? I am mostly discrete, however, I am confused by the MLP modules as inputs to Categorical distributions