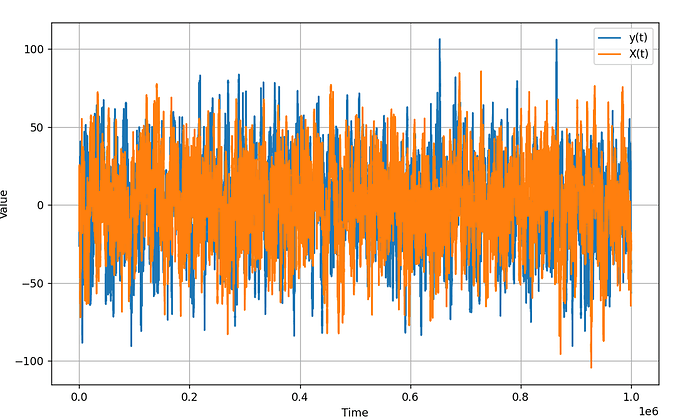
Hey guys - apologies for all the posts, but I am again running into some strange behavior with the GaussianHMM. I noticed some odd results that i was seeing, and ran this simulation - a simple 2-d random walk GaussianHMM:
import numpy as np
import matplotlib.pyplot as plt
import torch
import pyro
import pyro.distributions as dist
from torch.distributions import constraints
from pyro import poutine
pyro.set_rng_seed(1)
obs = torch.eye(2)
init_dist = dist.Normal(torch.zeros(2),1.).to_event(1)
transition = torch.eye(2)
transition_dist = dist.Normal(torch.zeros(2),1).to_event(1)
obs_dist = dist.Normal(torch.zeros(2),.01).to_event(1)
distt = dist.GaussianHMM(init_dist, transition, transition_dist, obs, obs_dist, duration=1000000)
sample = distt.rsample().detach()
x =sample[:,0]
y=sample[:,1]
t = np.linspace(1, 1000000, 1000000)
plt.figure(figsize=(10, 6))
plt.plot(t, y, label='y(t)')
plt.plot(t, x, label='X(t)')
plt.xlabel('Time')
plt.ylabel('Value')
plt.legend()
plt.grid(True)
plt.show()
Again, this is a simple 2-d random walk (shown as y and X in the image). After running 1 million steps, I got the following graph, which is clearly not a random walk, but rather looks stationary (see image below). This makes me think there is a bug in the distribution.