Dear fellow Pyro-maniacs,
I’m trying to make my model parallelizable in order to use Trace_ELBO(num_particles=100, vectorize_particles=True), and also Predictive(model, guide=guide, num_samples=100, parallel=True) (here 100 is arbitrary). I’ve read several tutorials on parallelizing code, but I can’t seem to make sense of it. So if anyone could help out, that would be really great. I’ve made a very simple version of the model: it’s just a GMM with 3 components on \mathbb{R}^2.
First I import some modules and define the GMM. It has fixed covariance matrices, and samples component weights and means (locations).
import pyro
import pyro.distributions as dist
from pyro.infer import Trace_ELBO, SVI, Predictive
from pyro.optim import Adam
from pyro.infer.autoguide import AutoMultivariateNormal, init_to_sample
import torch
import numpy as np
import scipy.stats as sts
import matplotlib.pyplot as plt
from tqdm import trange
def model(x, num_comp):
x_dim = x.shape[-1]
num_batch = x.shape[-2]
with pyro.plate("components", num_comp):
locs = pyro.sample(
"locs",
dist.Normal(
torch.zeros([x_dim]),
torch.ones([x_dim])
).to_event(1)
)
scales = 0.5*torch.ones((num_comp, x_dim))
alpha = torch.ones(num_comp)
weights = pyro.sample("weights", dist.Dirichlet(alpha))
with pyro.plate("data", num_batch):
d_mix = dist.Categorical(weights)
d_comp = dist.Normal(locs, scales).to_event(1)
pyro.sample("x", dist.MixtureSameFamily(d_mix, d_comp), obs=x)
guide = AutoMultivariateNormal(model, init_loc_fn=init_to_sample)
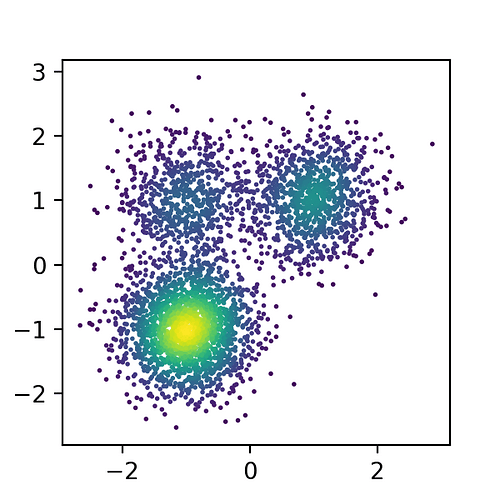
Now I generate some data.
N1 = 2000
N2 = 1000
N3 = 700
x1s = np.array([sts.norm.rvs(-np.ones(2), 0.5*np.ones(2)) for _ in range(N1)])
x2s = np.array([sts.norm.rvs(np.ones(2), 0.5*np.ones(2)) for _ in range(N2)])
x3s = np.array([sts.norm.rvs(np.array([-1,1]), 0.5*np.ones(2)) for _ in range(N3)])
xs = np.concatenate([x1s, x2s, x3s])
fig, ax = plt.subplots(1,1, figsize=(3,3))
dens = sts.gaussian_kde(xs.T)(xs.T)
ax.scatter(xs[:,0], xs[:,1], s=1, c=dens)
And this is the result.
Now I’m going to fit the model. Note that I’m setting vectorize_particles=False. It this is set to True, all hell breaks loose.
optim = Adam({"lr" : 0.1})
loss = Trace_ELBO(num_particles=100, vectorize_particles=False)
svi = SVI(model, guide, optim, loss)
# fit pyro model
epochs = 300
xs_tensor = torch.tensor(xs)
losses = []
for epoch in trange(epochs):
loss = svi.step(xs_tensor, 3)
losses.append(loss)
fig, ax = plt.subplots(1, 1, figsize=(4,3))
ax.plot(losses)
I won’t show the ELBO trace, but it looks good. Now I want to sample from the posterior using the Predictive class. I set parallel=False. Setting this to True causes errors.
pred = Predictive(model, guide=guide, num_samples=100, parallel=False)
sams = pred(xs_tensor, 3)
locs_sam = sams["locs"]
# show the sampled location parameters in relation to the data
fig, ax = plt.subplots(1, 1, figsize=(5,5))
ax.scatter(xs[:,0], xs[:,1], s=1, c=dens, alpha=0.3)
for i in range(3):
ax.scatter(locs_sam[:,i,0], locs_sam[:,i,1], s=5)
My question is: how do I change the model such that setting vectorize_particles=True and parallel=True does work? And also: what is the rationale behind this?
Thanks!