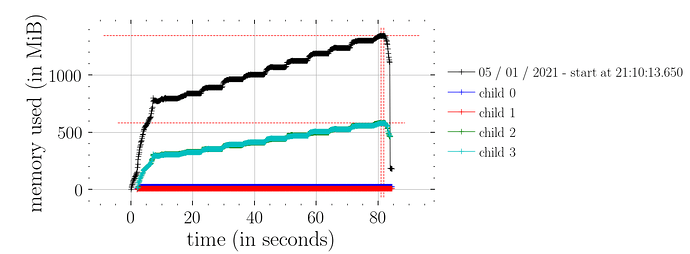
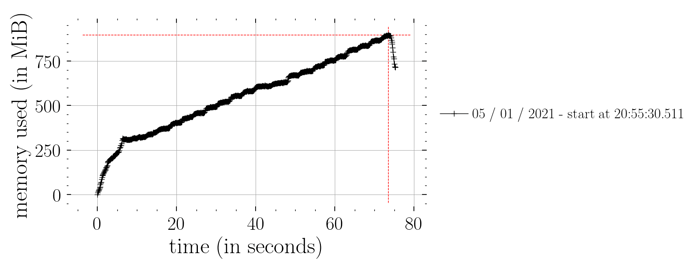
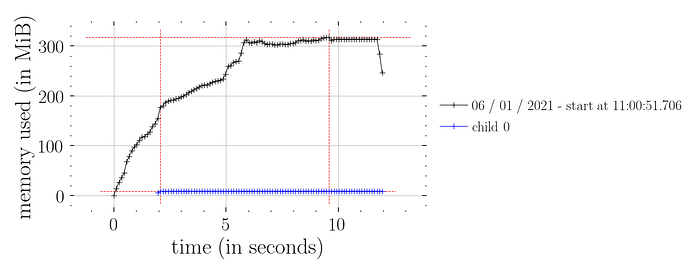
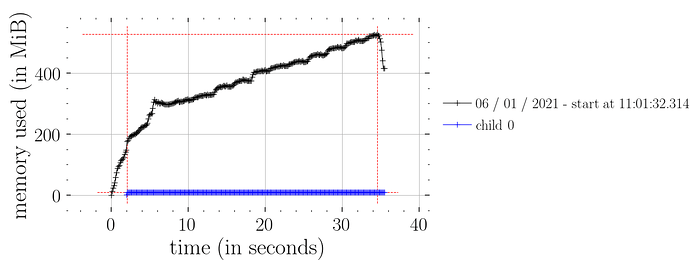
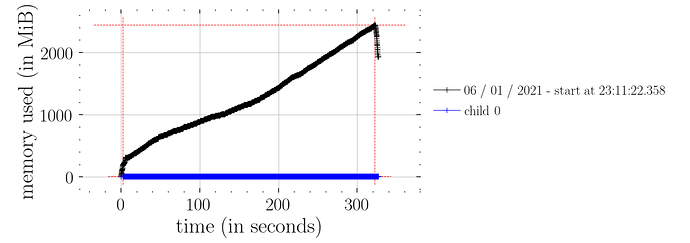
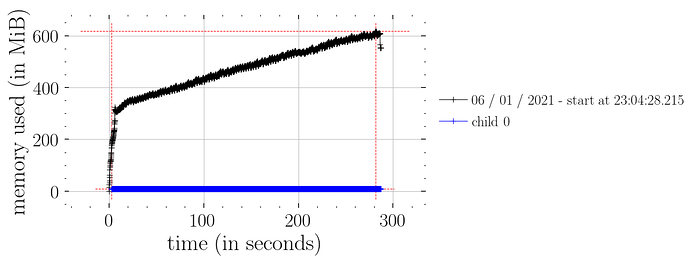
Hm, I tried to compare Predictive with the effect handler function defined in the Bayesian Regression Tutorial. When using the effect handler function, the memory consumption is constant in time compared to the one using Predictive. The rest of code below can be seen at the bottom of this post.
This works (i.e. constant time memory consumption):
N_runs = 100
for i in tqdm(range(N_runs)):
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, marriage=marriage, divorce=divorce)
samples_1 = mcmc.get_samples()
predict_fn = vmap(lambda rng_key, samples: predict(rng_key, samples, model_marriage, marriage=marriage))
predictions_1 = predict_fn(random.split(rng_key_, num_samples), samples_1)
whereas this doesn’t (i.e. not constant time memory consumption):
N_runs = 100
for i in tqdm(range(N_runs)):
rng_key, rng_key_ = random.split(rng_key)
mcmc.run(rng_key_, marriage=marriage, divorce=divorce)
samples_1 = mcmc.get_samples()
predictions = Predictive(model_marriage, samples_1)(rng_key_, marriage=marriage)["obs"]
I assume this is not as expected? Also, it seems that the effect handlers are a lot faster than Predictive? Timeit for Predictive was 939 ms ± 44.1 ms compared to 13.8 ms ± 1.6 ms for the effect handlers.
So I could, for now, just use the effect handlers method, however, I am having a bit of trouble getting it to work with my previous datasets mentioned. If I try:
predict_fn = vmap(lambda rng_key, samples: predict(rng_key, samples, model, data["z"], data["N"]))
predictions_1 = predict_fn(random.split(rng_key_, num_samples), samples_1)
where model and samples_1 are from my original dataset, it just runs and runs without getting anywhere, using quite a lot of cpu. I thought I would be able to just use this method on my own dataset as well. Do you happen to see what I miss here? If only this small “hack” could work, I would be more than happy to help with anything related to fixing Predictive if I can 
A last note, in the tutorial, in cell [12], where it says:
df['Mean Predictions'] = jnp.mean(predictions, axis=0)
shouldn’t it be:
df['Mean Predictions'] = jnp.mean(predictions_1, axis=0)
to reflect the new predictions from the effect handlers?
*Rest of the code:
def get_data_marriage():
DATASET_URL = (
"https://raw.githubusercontent.com/rmcelreath/rethinking/master/data/WaffleDivorce.csv"
)
dset = pd.read_csv(DATASET_URL, sep=";")
standardize = lambda x: (x - x.mean()) / x.std()
dset["AgeScaled"] = dset.MedianAgeMarriage.pipe(standardize)
dset["MarriageScaled"] = dset.Marriage.pipe(standardize)
dset["DivorceScaled"] = dset.Divorce.pipe(standardize)
return dset.MarriageScaled.values, dset.DivorceScaled.values
def model_marriage(marriage, divorce=None):
a = numpyro.sample("a", dist.Normal(0.0, 0.2))
bM = numpyro.sample("bM", dist.Normal(0.0, 0.5))
sigma = numpyro.sample("sigma", dist.Exponential(1.0))
mu = a + bM * marriage
numpyro.sample("obs", dist.Normal(mu, sigma), obs=divorce)
def predict(rng_key, post_samples, model, *args, **kwargs):
model = handlers.condition(handlers.seed(model, rng_key), post_samples)
model_trace = handlers.trace(model).get_trace(*args, **kwargs)
return model_trace["obs"]["value"]
rng_key = PRNGKey(0)
num_warmup, num_samples = 1000, 2000
marriage, divorce = get_data_marriage()
mcmc = MCMC(
NUTS(model_marriage),
progress_bar=False,
jit_model_args=True,
num_warmup=num_warmup,
num_samples=num_samples,
num_chains=1,
chain_method="sequential",
)
 )
)