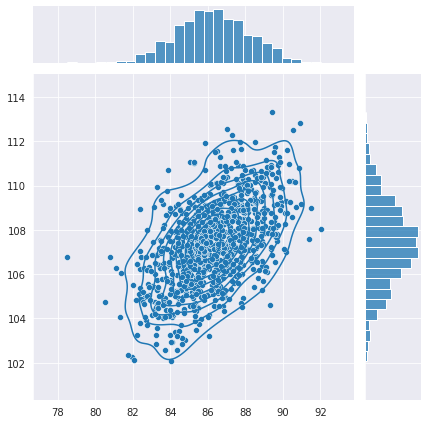
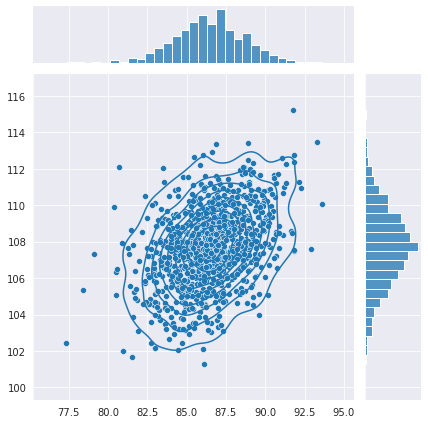
So, I am trying to create two regression models for multivariate outputs (2 outputs to be exact). The first model will explicitly infer the correlation while the second model I am specifying identity covariance matrix which I assumed would make the model act as if it is really just doing two independent linear regressions. Here is the code for the first model:
def model(x, y=None):
β = pyro.sample("β", dist.Normal(torch.zeros(x.shape[1],2),torch.ones(x.shape[1],2)).to_event(2))
B0 = pyro.sample("B0", dist.Normal(torch.zeros(2,),torch.ones(2,)))
μ = torch.matmul(x,β) + B0
θ = pyro.sample("θ", dist.HalfNormal(torch.ones(2)))
η = pyro.sample("η", dist.HalfNormal(1.))
L = pyro.sample("L", dist.LKJCorrCholesky(2, η))
L_Ω = torch.mm(torch.diag(θ.sqrt()), L)
with pyro.plate("data", x.shape[0]):
obs = pyro.sample("obs", dist.MultivariateNormal(loc=μ, scale_tril=L_Ω), obs=y)
And for the second model:
def model_no_cov(x, y=None):
β = pyro.sample("β", dist.Normal(torch.zeros(x.shape[1],2),torch.ones(x.shape[1],2)).to_event(2))
B0 = pyro.sample("B0", dist.Normal(torch.zeros(2,),torch.ones(2,)))
μ = torch.matmul(x,β) + B0
with pyro.plate("data", x.shape[0]):
obs = pyro.sample("obs", dist.MultivariateNormal(loc=μ, covariance_matrix=torch.eye(2)), obs=y)
What’s really driving me crazy is that the second model is producing outputs that are even more correlated than the actual data which you can see here:
I also should point out the second model has lower prediction error on the test set than the first model, which sort of boggles my mind. I’m not sure if I’m doing something wrong or my model just doesn’t make sense.