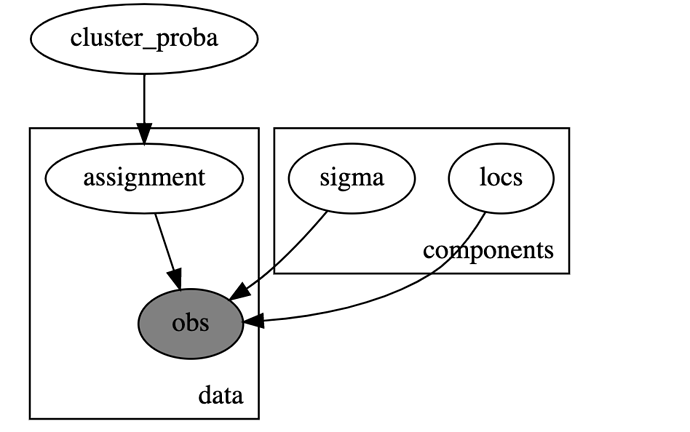
Hi everyone,
I am new to Pyro and I wanted to implement a multivariate gaussian mixture model.
The model runs without any errors, but when applied to simple and well separated data, it fails to infer the true parameters.
I used the following model in Numpyro:
def model(K,dim,data=None):
cluster_proba = numpyro.sample('cluster_proba',dist.Dirichlet(0.5 * jnp.ones(K)))
with numpyro.plate('components', K):
locs=numpyro.sample('locs',dist.MultivariateNormal(jnp.zeros(dim),10*jnp.eye(dim)))
sigma = numpyro.sample('sigma', dist.HalfCauchy(scale=10))
with numpyro.plate('data', len(data)):
assignment = numpyro.sample('assignment', dist.Categorical(cluster_proba),infer={"enumerate": "parallel"})
numpyro.sample('obs', dist.MultivariateNormal(locs[assignment,:][1], sigma[assignment][1]*jnp.eye(dim)), obs=data)
I used NUTS do to do the inference:
rng_key = jax.random.PRNGKey(0)
num_warmup, num_samples = 1000, 5000
kernel = NUTS(model)
mcmc = MCMC(
kernel,
num_warmup=num_warmup,
num_samples=num_samples,
)
mcmc.run(rng_key, data=data,K=3,dim=3)
mcmc.print_summary()
posterior_samples = mcmc.get_samples()
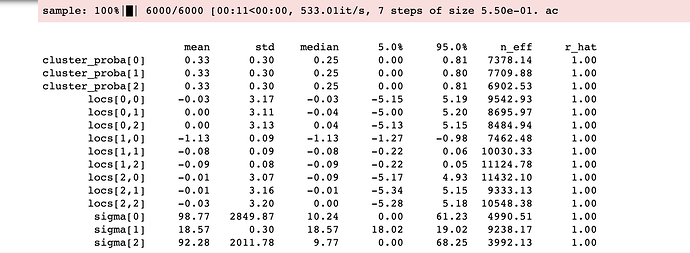
Even though the model runs, it seems that it doesn’t manage to converge to correct values when applied to well separated (simulated data). In particular the weights are not updated and remain at 1/3 (when applied to three components)
I simulated 3D data coming from 3 components using the code below:
n = 2500 # Total number of samples
k = 3 # Number of clusters
dim=3 # Number of dimension
p_real = np.array([0.1, 0.5, 0.4]) # Probability of choosing each cluster
mu0=[10, 10, 10]
mu1=[-5, -3, -3]
mu2=[1, 1, 1]
mus=[mu0,mu1,mu2]
sigma0=1
sigma1=0.5
sigma2=0.5
sigmas=[sigma0,sigma1,sigma2]
clusters = np.random.choice(k, size=1, p=p_real)
data=np.random.multivariate_normal(mus[clusters[0]],sigmas[clusters[0]]*np.eye(dim), (1))
for i in range(1,n):
clusters = np.random.choice(k, size=1, p=p_real)
mu=mus[clusters[0]]
sigma=sigmas[clusters[0]]*np.eye(dim)
data_point=np.random.multivariate_normal(mu, sigma,(1))
data=np.concatenate((data,data_point), axis=0)
data
This gives the results below that are incoherent with the data provided:
If anyone can tell me what I am doing wrong that would be much appreciated.
Thanks!