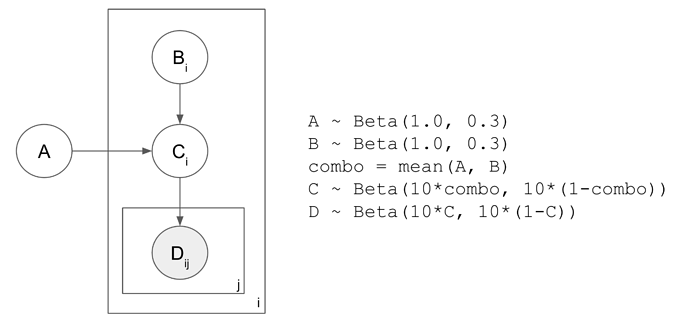
Hi, back again with a nan_mask question. This time I’m getting unexpected SVI behavior from instances which have all of their descendant observations nan’d out.
Toy model is below. Everything is drawn from a Beta distribution. C tracks the mean of A and B; D tracks C.
The issue in the simplest case is when i=2, but all D_1j are observed as nan, e.g.
D = [[0.9, 0.9, 0.9, ..., 0.9], [nan, nan, nan, ... nan]]. In that case A, B_0, and C_0 have descendant observations at 0.9, but B_1 and C_1 only have nan descendant observations. C_1 still has some indirect connection to the non-nan observations via C_1 <- A -> C_0 -> D_0j.
B_1, however, is d-separated due to the v-structure and therefore I would expect SVI to just draw it from the prior. I’ve confirmed this is the case when A is removed from the model. But when A is present the prediction for B_1 clearly deviates from the prior mean of 0.75, as below. Note that this happens in the opposite direction if the prior is flipped to Beta(0.3, 1.0): then B_1 deviates below the prior mean of 0.25. And if the prior is uniform at Beta(1.0, 1.0) then B_1 stays around the expectation of 0.5…
A: [0.8]
B: [0.89, 0.89]
C: [0.86, 0.84]
Any idea why? The real model is much larger and more complex, and it would be much better to not have to worry about filtering things out that may be missing all descendant observations, and instead just be able to assume any such predictions are drawn from the prior.
MWE
Note: making sure to properly handle nan observations as fixed in my previous post.
import pyro
import torch
import pyro.distributions as dist
import torch.distributions.constraints as constraints
from typing import List
num_BC = 2
num_D = 100
def model(obs: torch.Tensor, nan_mask: torch.Tensor):
A = pyro.sample('A', dist.Beta(1, .3))
with pyro.plate('BC_plate', size=num_BC, dim=-1):
B = pyro.sample('B', dist.Beta(1, .3))
parent_values = torch.stack((B, A.expand(B.shape)))
combined_parent_values = torch.mean(parent_values, dim=0)
C = pyro.sample('C', dist.Beta(10 * combined_parent_values, 10 * (1 - combined_parent_values)))
with pyro.plate('D_plate', size=num_D, dim=-2):
C_expanded = C.expand(num_D, num_BC)
with pyro.poutine.mask(mask=nan_mask):
D = pyro.sample('D', dist.Beta(10 * C_expanded, 10 * (1 - C_expanded)), obs=obs)
def guide(obs: torch.Tensor, nan_mask: torch.Tensor):
A_concentrations = get_concentration_params('A', 1)
pyro.sample('A', dist.Beta(*A_concentrations))
with pyro.plate('BC_plate', size=num_BC, dim=-1):
B_concentrations = get_concentration_params('B', num_BC)
pyro.sample('B', dist.Beta(*B_concentrations))
C_concentrations = get_concentration_params('C', num_BC)
pyro.sample('C', dist.Beta(*C_concentrations))
def get_concentration_params(site_name: str, size: int) -> List[pyro.param]:
# Beta distribution concentration1 and concentration0 parameters
return [
pyro.param(f'{site_name}_concentration{i}', torch.ones(size), constraint=constraints.greater_than(0.1))
for i in reversed(range(2))
]
def main():
obs = torch.full((num_D, num_BC), 0.9)
obs[:, -1] = float('nan')
nan_mask = ~torch.isnan(obs)
obs[torch.isnan(obs)] = 0.4242 # to avoid nan loss
svi = pyro.infer.SVI(
model,
guide,
pyro.optim.Adam({"lr": 0.1, "betas": (0.95, 0.999)}),
loss=pyro.infer.Trace_ELBO(),
)
for _ in range(10000):
svi.step(obs, nan_mask)
param_store = pyro.get_param_store()
for site_id in ['A', 'B', 'C']:
concentration1 = param_store[f'{site_id}_concentration1']
concentration0 = param_store[f'{site_id}_concentration0']
beta_mean = concentration1 / (concentration1 + concentration0)
print(f'{site_id}: ', [round(float(v), 2) for v in beta_mean])
if __name__ == '__main__':
main()