Hi, I am new to pyro.
I am following your tutorial: https://pyro.ai/examples/lda.html, which is very clear and easy to read.
I want to apply it on binary matrix, where “num-of-words-per-document” is fixed and the observations (word) are drawn from beta-bernoulli distribution instead of dirichlet-categorical distribution.
Let K to be the number of topics, S to be the volumne of vocabulary, L to be the number of words in each document. In the simplest case, L is fixed and L=S.
Then,
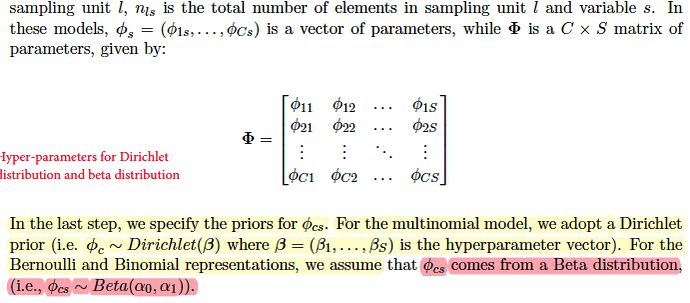
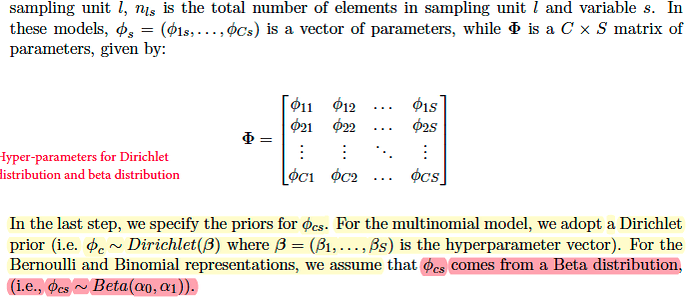
- for the original case where each word is drawn from Categorical Distribution, topic-word prior has K parameterss β. The length of each β is S. So the shape of parameter B=[β1,β2,…, βk] is (K, S).
- for the case of binary observations where each entry is drawn from Bernoulli Distribution, topic -word prior should have K *S pairs of Beta parameters (α0,α1). Referring to the paper Rlda, each φcs for each entry in the binary case is drawn from a beta distribution while φc for each cluster in the multinomial case is drawn from dirichlet distrbution.
So I modify your codes to be like this (only the for generative part):
def model(data=None, args=None, batch_size=None):
# Globals.
with pyro.plate(“topics”, args.num_topics):
topic_weights = pyro.sample(“topic_weights”, dist.Gamma(1. / args.num_topics, 1.))
print(“topic weights”)
print(topic_weights.shape)
# NOTE: phi prior
# topic_words = pyro.sample(“topic_words”,
# dist.Dirichlet(torch.ones(args.num_words) / args.num_words))
topic_words = pyro.sample(“topic_words”, dist.Beta(
torch.ones(args.num_words)*0.5, torch.ones(args.num_words)*0.5)
)
print(“topic words”)
print(topic_words.shape)
# Locals.
with pyro.plate("documents", args.num_docs) as ind:
if data is not None:
with pyro.util.ignore_jit_warnings():
assert data.shape == (args.num_words_per_doc, args.num_docs)
data = data[:, ind]
doc_topics = pyro.sample("doc_topics", dist.Dirichlet(topic_weights))
with pyro.plate("words", args.num_words_per_doc):
# The word_topics variable is marginalized out during inference,
# achieved by specifying infer={"enumerate": "parallel"} and using
# TraceEnum_ELBO for inference. Thus we can ignore this variable in
# the guide.
word_topics = pyro.sample("word_topics", dist.Categorical(doc_topics),
infer={"enumerate": "parallel"})
# NOTE: phi prior
# data = pyro.sample("doc_words", dist.Categorical(topic_words[word_topics]),
# obs=data)
data = pyro.sample("doc_words", dist.Bernoulli(topic_words[word_topics]),
obs=data)
return topic_weights, topic_words, data
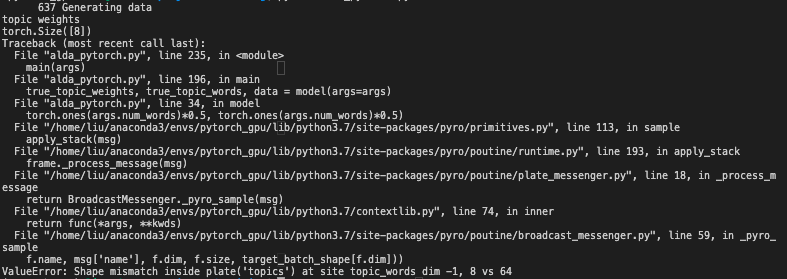
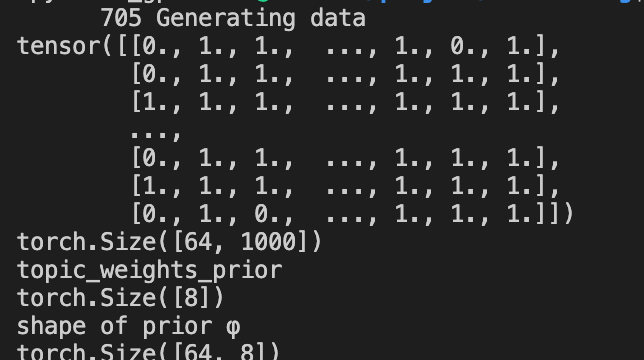
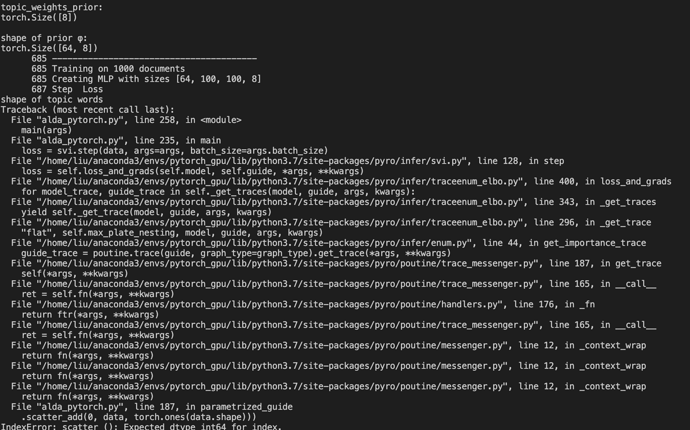
I tested this part of codes but got shape mismatch problem (# of topics=8, # of words=64, # of documents=1000):
Could you help me figure out the error I’ve made? Thanks very much!