Hi,
I’m trying a very simple exercise in Pyro – to learn the parameters of a univariate normal distribution via MLE. I’m doing this for purely learning purposes. While the MLE is trivial (dataset mean and dataset variance), I am trying to do the same using autograd. I see that there are examples on estimating the full Bayesian via SVI, however, I am unaware of examples on the trivial MLE.
import pyro
import pyro.distributions as dist
uv_normal = dist.Normal(loc=1., scale=1.)
train_data = uv_normal.sample([10000])
Once I have the samples, I create a loc parameter.
pyro.clear_param_store()
loc = pyro.param("loc", init_tensor=torch.tensor(0.1))
and then use simple gradient descent where I compute the gradient of the negative log likelihood (-torch.mean(to_train.log_prob(train_data)) to update loc.
for i in range(10):
to_train = dist.Normal(loc=loc, scale=1.0)
loss = -torch.mean(to_train.log_prob(train_data))
loss.backward()
print(to_train, loss, loc, loc.grad)
loc = loc - 0.00001*loc.grad
However, I get the output and the error below:
Normal(loc: 0.10000000149011612, scale: 1.0) tensor(1.4146, grad_fn=<NegBackward0>) tensor(0.1000, requires_grad=True) tensor(0.0800)
Normal(loc: 0.09999920427799225, scale: 1.0) tensor(1.4146, grad_fn=<NegBackward0>) tensor(0.1000, grad_fn=<SubBackward0>) None
TypeError Traceback (most recent call last)
Input In [99], in <module>
7 loss.backward()
8 print(to_train, loss, loc, loc.grad)
----> 9 loc = loc - 0.00001*loc.grad
TypeError: unsupported operand type(s) for *: 'float' and 'NoneType'
Clearly, the gradient becomes None.
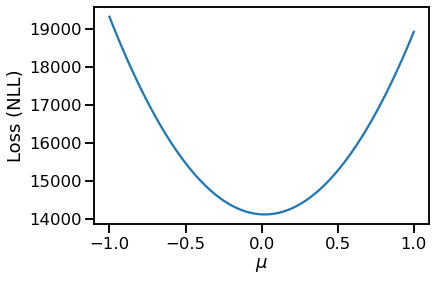
Ofcourse, if I visually inspect the loss (NLL) for a discrete set of locs, I get the expected plot:
losses = {}
for i in torch.linspace(-1.0, 1.0, 100):
losses[i.item()] = -torch.sum(dist.Normal(loc=i, scale=1.0).log_prob(train_data)).item()
import pandas as pd
pd.Series(losses).plot(xlabel=r"$\mu$", ylabel="Loss (NLL)")
However, I was able to use pretty much the same code as above in TF Probability as follows to learn the MLE for loc.
Working TensorFlow Probability code for the same problem
import tensorflow as tf
import tensorflow_probability as tfp
uv_normal = tfd.Normal(loc=0., scale=1.)
train_data = uv_normal.sample(10000)
to_train = tfd.Normal(loc = tf.Variable(-1., name='loc'), scale = 1.)
def nll(train):
return -tf.reduce_mean(to_train.log_prob(train))
def get_loss_and_grads(train):
with tf.GradientTape() as tape:
tape.watch(to_train.trainable_variables)
loss = nll(train)
grads = tape.gradient(loss, to_train.trainable_variables)
return loss, grads
optimizer = tf.keras.optimizers.Adam(learning_rate=0.01)
iterations = 500
losses = np.empty(iterations)
vals = np.empty(iterations)
for i in range(iterations):
loss, grads = get_loss_and_grads(train_data)
losses[i] = loss
vals[i] = to_train.trainable_variables[0].numpy()
optimizer.apply_gradients(zip(grads, to_train.trainable_variables))
if i%50 == 0:
print(i, loss.numpy())
After the 500 iterations, I am able to learn the correct loc in TFP.
- I wanted to check if I am making a mistake or missing something obvious. Or, in short, what would be the best way to compute the MLE for the above problem for estimating the parameters of the distribution?
- What would be an efficient way to write the following code:
for i in torch.linspace(-1.0, 1.0, 100):
losses[i.item()] = -torch.sum(dist.Normal(loc=i, scale=1.0).log_prob(train_data)).item()
If I’m able to get this to work, I’d be happy to make PRs to the documentation and add examples on MLE, MAP, and Full Bayesian (via SVI). As an example, I created a post on Coin tosses: MLE, MAP and Full Bayesian using TFP. I’d like to be able to get it working on Pyro.