Hi there,
So I have a little toy Gaussian Mixture Model that I’ve been trying to get to work with mini-batching. The problem is that while the model appears to work perfectly when passing in the whole dataset, attempting to minibatch totally messes up the training. I’m wondering if there’s something I don’t understand about minibatching or indexing global sites that is causing the issue? I’ve read all the tutorials and the non-amortized minibatching forum posts and I can’t seem to find a good example.
Setup:
import os
from collections import defaultdict
import torch
import numpy as np
import scipy.stats
from torch.distributions import constraints
from matplotlib import pyplot
import seaborn
%matplotlib inline
import pyro
import pyro.distributions as dist
from pyro import poutine
from pyro.infer.autoguide import AutoDelta
from pyro.optim import Adam
from pyro.infer import SVI, TraceEnum_ELBO, config_enumerate, infer_discrete
smoke_test = ('CI' in os.environ)
assert pyro.__version__.startswith('1.7.0')
from pyro.distributions import MultivariateNormal
#synthetic 4 gaussian dataset in 2 dimensions
data = torch.cat((MultivariateNormal(-8 * torch.ones(2), torch.eye(2)).sample([50]),
MultivariateNormal(8 * torch.ones(2), torch.eye(2)).sample([50]),
MultivariateNormal(torch.tensor([1.5, 2]), torch.eye(2)).sample([50]),
MultivariateNormal(torch.tensor([-0.5, 1]), torch.eye(2)).sample([50])))
So the original model can be run as follows:
K = 4 # Fixed number of components.
n_var=2
n_obs=data.shape[0]
@config_enumerate
def model(data):
# Global variables.
with pyro.plate('data', n_obs):
weights = pyro.sample('weights', dist.Dirichlet(0.5 * torch.ones(K)))
with pyro.plate('vars',n_var):
with pyro.plate('components', K):
locs = pyro.sample('locs', dist.Normal(0., 10.))
scale = pyro.sample('scale', dist.LogNormal(0., 2.))
#this doesn't actually minibatch, just makes analogous to minibatching scheme below
with pyro.plate('minibatch', n_obs):
# Local variables.
assignment = pyro.sample('assignment', dist.Categorical(weights))
pyro.sample('obs', dist.Normal(locs[assignment,:], scale[assignment,:]).to_event(1), obs=data)
optim = pyro.optim.ClippedAdam({'lr': 0.1})
elbo = TraceEnum_ELBO(max_plate_nesting=2)
def init_loc_fn(site):
if site["name"] == "weights":
# Initialize weights to uniform.
return torch.ones((n_obs,K)) / K
if site["name"] == "scale":
return (data.var(0) / 2).sqrt().expand(K,n_var)
if site["name"] == "locs":
return data[torch.multinomial(torch.ones(n_obs) / n_obs, K),:]
raise ValueError(site["name"])
def initialize(seed):
global global_guide, svi
pyro.set_rng_seed(seed)
pyro.clear_param_store()
global_guide = AutoDelta(poutine.block(model, expose=['weights', 'locs', 'scale']),
init_loc_fn=init_loc_fn)
svi = SVI(model, global_guide, optim, loss=elbo)
return svi.loss(model, global_guide, data)
# Choose the best among 100 random initializations.
loss, seed = min((initialize(seed), seed) for seed in range(100))
initialize(seed)
print('seed = {}, initial_loss = {}'.format(seed, loss))
# Register hooks to monitor gradient norms.
gradient_norms = defaultdict(list)
for name, value in pyro.get_param_store().named_parameters():
value.register_hook(lambda g, name=name: gradient_norms[name].append(g.norm().item()))
losses = []
for i in range(200 if not smoke_test else 2):
loss > = svi.step(data)
losses.append(loss)
print('.' if i % 100 else '\n', end='')
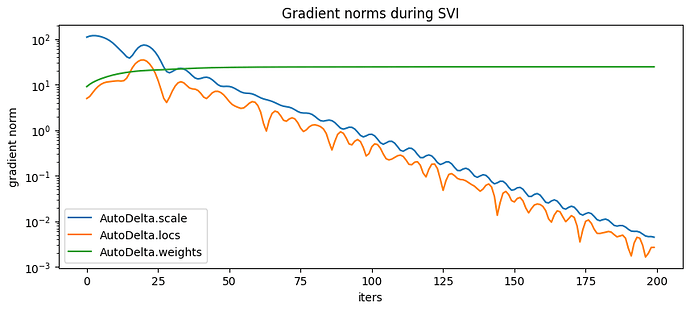
pyplot.figure(figsize=(10,4), dpi=100).set_facecolor('white')
for name, grad_norms in gradient_norms.items():
pyplot.plot(grad_norms, label=name)
pyplot.xlabel('iters')
pyplot.ylabel('gradient norm')
pyplot.yscale('log')
pyplot.legend(loc='best')
pyplot.title('Gradient norms during SVI')
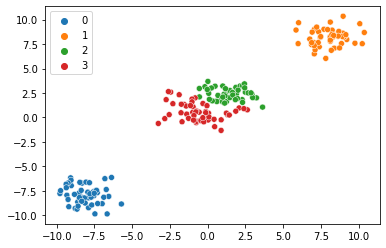
map_estimates = global_guide(data)
weights = map_estimates['weights']
seaborn.scatterplot(x=data[:,0].numpy(),y=data[:,1].numpy(), hue=map_estimates['weights'].max(1).indices.cpu().detach().numpy().astype(str))
Minibatching Model Fails
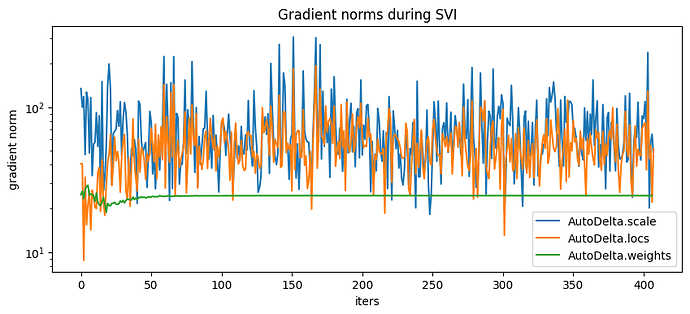
So now if I make slight modifications to the model and training, such that the weights have global parameters but the categorical and ELBO evaluation are minibatched, the model is garbage.
Dataloader that also returns indices for global variables.
subsample_size=32
from torch.utils.data import TensorDataset, DataLoader
def dataset_with_indices(cls):
"""
Modifies the given Dataset class to return a tuple data, target, index
instead of just data, target.
"""
def __getitem__(self, index):
data = cls.__getitem__(self, index)
return data, index
return type(cls.__name__, (cls,), {
'__getitem__': __getitem__,
})
#d=TensorDataset(data)
IndexedTensorDataset=dataset_with_indices(TensorDataset)
d=IndexedTensorDataset(data)
dataloader = DataLoader(d, batch_size=subsample_size, shuffle=True,drop_last=True)
Model and Training
K = 4 # Fixed number of components.
n_var=2
n_obs=data.shape[0]
@config_enumerate
def model(data_batch):
data=data_batch[0][0]
# Global variables.
with pyro.plate('data', n_obs):
weights = pyro.sample('weights', dist.Dirichlet(0.5 * torch.ones(K)))
with pyro.plate('vars',n_var):
with pyro.plate('components', K):
locs = pyro.sample('locs', dist.Normal(0., 10.))
scale = pyro.sample('scale', dist.LogNormal(0., 2.))
with pyro.plate('minibatch', n_obs,subsample=data_batch[1].long()) as ind:
# Local variables.
assignment = pyro.sample('assignment', dist.Categorical(weights[ind,:]))
pyro.sample('obs', dist.Normal(locs[assignment,:], scale[assignment,:]).to_event(1), obs=data)
#optimization setup same as above
optim = pyro.optim.ClippedAdam({'lr': 0.1})
elbo = TraceEnum_ELBO(max_plate_nesting=2)
def init_loc_fn(site):
if site["name"] == "weights":
# Initialize weights to uniform.
return torch.ones((n_obs,K)) / K
if site["name"] == "scale":
return (data.var(0) / 2).sqrt().expand(K,n_var)
if site["name"] == "locs":
return data[torch.multinomial(torch.ones(n_obs) / n_obs, K),:]
raise ValueError(site["name"])
def initialize(seed):
global global_guide, svi
pyro.set_rng_seed(seed)
pyro.clear_param_store()
global_guide = AutoDelta(poutine.block(model, expose=['weights', 'locs', 'scale']),
init_loc_fn=init_loc_fn)
svi = SVI(model, global_guide, optim, loss=elbo)
return svi.loss(model, global_guide, data)
# Choose the best among 100 random initializations.
loss, seed = min((initialize(seed), seed) for seed in range(100))
initialize(seed)
print('seed = {}, initial_loss = {}'.format(seed, loss))
# Register hooks to monitor gradient norms.
gradient_norms = defaultdict(list)
for name, value in pyro.get_param_store().named_parameters():
value.register_hook(lambda g, name=name: gradient_norms[name].append(g.norm().item()))
losses = []
for i in range(200 if not smoke_test else 2):
for data_batch in iter(dataloader):
loss = svi.step(data_batch)
losses.append(loss)
print('.' if i % 100 else '\n', end='')
pyplot.figure(figsize=(10,4), dpi=100).set_facecolor('white')
for name, grad_norms in gradient_norms.items():
pyplot.plot(grad_norms, label=name)
pyplot.xlabel('iters')
pyplot.ylabel('gradient norm')
pyplot.yscale('log')
pyplot.legend(loc='best')
pyplot.title('Gradient norms during SVI');
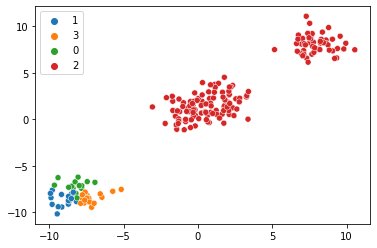
map_estimates = global_guide(data)
weights = map_estimates['weights']
import seaborn
seaborn.scatterplot(x=data[:,0].numpy(),y=data[:,1].numpy(), hue=map_estimates['weights'].max(1).indices.cpu().detach().numpy().astype(str))
Thanks and sorry for the very long post!
MT