Hi, I am trying to fit a Poisson regression model with MCMC sampling.
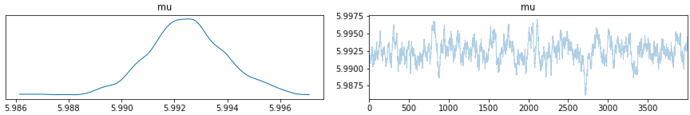
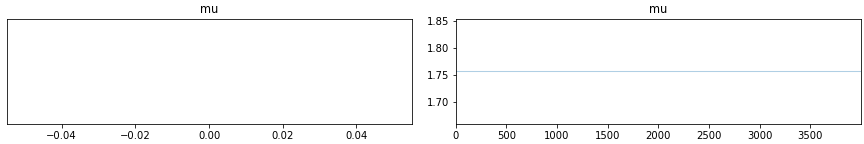
My problem is that the sampling breaks down for high values of the rate of the Poisson process. Take the minimal reproducer below: if I set the mean value of my (dummy) data to 400, everything is fine and I can fit the model (top plots below). However, if I increase the value to e.g. 1000, the sampling breaks down completely, see bottom plots below. Any idea what I am doing wrong?
import numpyro as nmp
from numpyro.infer import MCMC, NUTS, Predictive
import numpyro.distributions as dist
from jax import random
import jax.numpy as jnp
from numpy.random import default_rng
import arviz as az
# Create dummy data
rng = default_rng()
mean = 400
samples = rng.poisson(mean, size=1000)
# Define model
def model(data):
mu = nmp.sample('mu', dist.Normal(8., 2.))
lam = jnp.exp(mu)
y = nmp.sample("y", dist.Poisson(lam),
obs=data)
# Fit model
nuts_kernel = NUTS(model, target_accept_prob=0.99)
mcmc = MCMC(nuts_kernel, num_samples=4000, num_warmup=2000, num_chains=1)
rng_key = random.PRNGKey(0)
mcmc.run(rng_key, samples)
az.plot_trace(mcmc)