Hi there! I’m new to pyro and I’m trying out a toy example of Dirichlet Process Mixture Model, implemented with truncation. I chose the prior for each component to have its own mean and covariance parameter, which are independently drawn. My model is like this:
T = 10
def mix_weights(beta):
beta1m_cumprod = (1 - beta).cumprod(-1)
return F.pad(beta, (0, 1), value=1) * F.pad(beta1m_cumprod, (1, 0), value=1)
def model(data):
alpha = pyro.param("alpha", torch.tensor([1.0]))
with pyro.plate("sticks", T-1):
beta = pyro.sample("beta", Beta(1, alpha))
with pyro.plate("component", T):
mu = pyro.sample("mu", MultivariateNormal(torch.zeros(d), 5 * torch.eye(d)))
cov = pyro.sample("cov", Wishart(df=d, covariance_matrix=torch.eye(d)))
with pyro.plate("data", N):
z = pyro.sample("z", Categorical(mix_weights(beta)))
pyro.sample("obs", MultivariateNormal(mu[z], precision_matrix=cov[z]), obs=data)
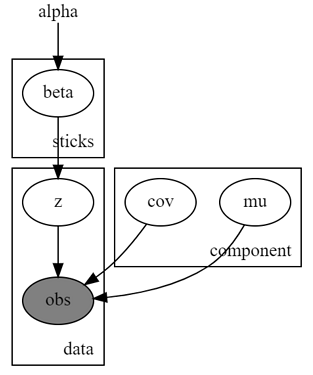
The graphical model may be more intuitive:
After this model construction, I tried to use MCMC sampling for posterior inference, with the following code:
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, num_samples=500, warmup_steps=100)
mcmc.run(data)
posterior_samples = mcmc.get_samples()
However, this raises an error, which I thought should be related to the Wishart components in my model.
NotImplementedError: Cannot transform _PositiveDefinite constraints
How can I avoid this error? Should I change the prior, or use other methods like Cholesky decomposition to ensure the constraints implicitly?
Thanks for any advice!