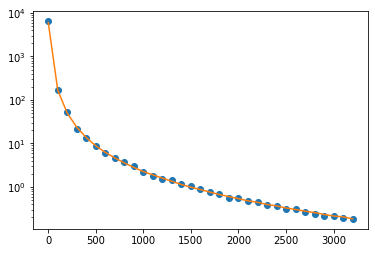
I’m pretty new to PPLs and have been exploring using Monte Carlo methods to fit some experimental physical data. The problem is the code in numpyro is super slow. I’m using a GPU from colab and it seems to spend abound 5 mins in background_compile() and _execute_compiled() before showing the progress bar. At this point it predicts it will take ~100 hours to complete the sampling for 1000 burn-in, 10000 steps and 250 vectorised chains. This seems quite long to me and I’m wondering if anyone with more experience of this believes this is typical or if my code is not optimal? In addition, the GPU memory pretty instantly reaches 13 GB of the 15 GB once running.
The code is below and hopefully well commented. To test everything I’ve simulated data from the model and have fixed all but one of the variables. The time scale is (0, 3200) ns with $\Delta$t = 100 ns. I have tried solving the ODEs with jax.experimental.ode.odeint but have the same problems.
Thanks in advance for any suggestions/insights. ![]()
![]()
#Import required packages
import jax
import jax.numpy as jnp
from jax.random import PRNGKey
from jax.experimental.ode import odeint
import numpyro
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS
import diffrax
import equinox as eqx
#Set all floats to 64 bit
jax.config.update("jax_enable_x64", True)
#Rate equations charge carrier dynamics
class BTDP(eqx.Module):
"""
Rate equations for the BTD model, as per doi.org/10.1039/d0cp04950f p. 28348
including trapping, depopulation and background doping.
Model allows for a non-constant density of traps with depopulation from these
traps to the valence band.
Returns
-------
[f0, f1, f2]: array
Array of the rate equations. Dynamics for electron, trap and hole
concentraions.
"""
kt: float
kb: float
kd: float
NT: float
p0: float
def __call__(self, t, y, args):
B = self.kb * y[0] * (y[2] + self.p0)
T = self.kt * y[0] * (self.NT- y[1])
D = self.kd * y[1] * (y[2] + self.p0)
f0 = -B - T
f1 = T - D
f2 = -B - D
return jnp.stack([f0, f1, f2])
#JIT compiled function to solve the ODE
@jax.jit
def solve_TRPL_BTDP(kt, kb, kd, NT, p0, N0):
"""
Solve the ODEs for the BTD model.
Parameters
----------
kt: float
k_T trapping rate constant (cm^3 ns^-1).
kb: float
k_B bimolecular rate constant (cm^3 ns^-1).
kd: float
k_D depopulation rate constant (cm^3 ns^-1) (trap to valence band).
NT: float
Trap density (cm^-3).
p0: float
Doping density (cm^-3).
N0: float
Initial electron concentration (cm^-3).
Returns
-------
sol: array
Solution to the ODEs.
"""
#Define equations
btdp = BTDP(kt, kb, kd, NT, p0)
terms = diffrax.ODETerm(btdp)
#Start and end times
t0 = 0.0
t1 = 3200.0
#Initial conditions and initial time step
y0 = jnp.array([N0, 0.0, N0 + p0])
dt0 = 0.0002
#Define solver and times to save at
solver = diffrax.Kvaerno5()
saveat = diffrax.SaveAt(ts=jnp.arange(33, dtype=jnp.float64)*100)
#Controller for adaptive time stepping
stepsize_controller = diffrax.PIDController(rtol=1e-8, atol=1e-8)
#Solve ODEs
sol = diffrax.diffeqsolve(
terms,
solver,
t0,
t1,
dt0,
y0,
saveat=saveat,
stepsize_controller=stepsize_controller,
)
return sol
#JIT compiled function to calculate the TRPL signal
@jax.jit
def TRPL_BTDP(kt, kb, kd, NT, p0, y0, N0):
"""
Calculate the TRPL signal for the BTD model.
Parameters
----------
kt: float
k_T trapping rate constant (cm^3 ns^-1).
kb: float
k_B bimolecular rate constant (cm^3 ns^-1).
kd: float
k_D depopulation rate constant (cm^3 ns^-1) (trap to valence band).
NT: float
Trap density (cm^-3).
p0: float
Doping density (cm^-3).
y0: float
Initial TRPL counts (counts).
N0: float
Initial electron concentration (cm^-3).
Returns
-------
sig: array
TRPL signal.
"""
#Solve ODEs
sol = solve_TRPL_BTDP(kt, kb, kd, NT, p0, N0)
#Calculate TRPL signal
sig = jnp.log10(sol.ys[:, 0]) + jnp.log10(sol.ys[:, 2])
#Normalise signal and correct for the initial TRPL counts
sig = sig - sig[0] + jnp.log10(y0)
return sig
#Standardise the data
def _standardise(x):
"""
Standardise the data to have a mean of 0 and a standard deviation of 1.
Parameters
----------
x: numpy.ndarray
The data to standardise.
Returns
-------
x: numpy.ndarray
The standardised data.
"""
return (x - x.mean()) / x.std()
#JIT compiled function to standardise the data
standardise = jax.jit(_standardise)
#warm up the JIT
standardise(TRPL_BTDP(1e-15, 1e-18, 0, 1e13, 2e13, 6000, 1e15)).block_until_ready()
#Bayesian model
def model(N0, y, y0):
"""
Bayesian model for the BTDP model.
Parameters
----------
N0: float
Initial electron concentration (cm^-3).
y: array
log10 of the experimental TRPL signal.
y0: float
Initial TRPL counts (counts).
"""
#Define priors for the rate constants in log space
theta = numpyro.sample(
"theta",
dist.TruncatedNormal(
low = jnp.array([-16]),
high = jnp.array([-14]),
loc = jnp.array([-15]),
scale = jnp.array([1]),
),
)
#Calculate the TRPL signal and standardise
#Testing with fixed parameters for all but kt
signal = TRPL_BTDP(10**theta[0], 5e-18, 0, 1e13, 2e13, y0, N0)
signal = standardise(signal)
#Define the likelihood
numpyro.sample("y", dist.Normal(signal, .02), obs=y)
#Generate some simulated data to test the fitting
#time points fixed in model for now
N0 = 1e16
y = TRPL_BTDP(2.2e-15, 5e-18, 0, 1e13, 2e13, 6000, N0)
# create a normal distribution object
normal = dist.Normal(0, .02)
# generate random samples from the normal distribution
noise = normal.sample(PRNGKey(84), (33,))
num_warmup, num_samples = 1000, 10000
#Define the MCMC
mcmc = MCMC(
NUTS(model, dense_mass=True),
num_warmup=num_warmup,
num_samples=num_samples,
num_chains=250,
chain_method="vectorized",
progress_bar=True,
)
#Run the MCMC with the simulated data and noise
mcmc.run(PRNGKey(78), N0 = N0, y = standardise(y+noise), y0 = 10**(y+noise)[0])
mcmc.print_summary()