Hi all,
I’m trying to implement Manifold Relevance Determination using pyro.
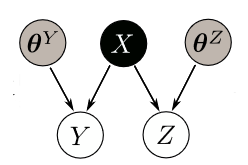
In this model, there are two visible variables (Y and Z) and one hidden variable (X). The hidden variable is mapped to visible domain using two variational sparse Gaussian processes. The model is equivalent to a GPLVM with multiple views.
I implemented the model as follows :
class MRD(Parameterized):
def __init__(self, base_model1,base_model2):
super().__init__()
if base_model1.X.dim() != 2 or base_model2.X.dim() != 2:
raise ValueError(
"GPLVM model only works with 2D latent X, but got "
"X.dim() = {}.".format(base_model1.X.dim())
)
self.base_model1 = base_model1
self.base_model2 = base_model2
self.X = PyroSample(dist.Normal(base_model1.X.new_zeros(base_model1.X.shape), 1.0).to_event())
self.autoguide("X", dist.Normal)
self.X_loc.data = base_model1.X
@pyro_method
def model(self):
self.mode = "model"
self.base_model1.set_data(self.X, self.base_model1.y)
self.base_model2.set_data(self.X, self.base_model2.y)
self.base_model1.model()
self.base_model2.model()
@pyro_method
def guide(self):
self.mode = "guide"
# self._load_pyro_samples()
self.base_model1.set_data(self.X, self.base_model1.y)
self.base_model2.set_data(self.X, self.base_model2.y)
self.base_model1.guide()
self.base_model2.guide()
def predict_base_model1(self, Xnew):
self.mode = "guide"
self.base_model1.set_data(self.X, self.base_model1.y)
return self.base_model1(Xnew)
def predict_base_model2(self, Xnew):
self.mode = "guide"
self.base_model2.set_data(self.X, self.base_model2.y)
return self.base_model2(Xnew)
num_latent=10
num_inducing=100
## VSGP
X_init = torch.zeros(x_train.shape[0], num_latent,device='cuda')
Xu = torch.zeros(num_inducing, num_latent)
likelihood1 = gp.likelihoods.Gaussian()
likelihood2 = gp.likelihoods.Gaussian()
kernel1 = gp.kernels.RBF(input_dim=num_latent, lengthscale=torch.ones(num_latent))
kernel2 = gp.kernels.RBF(input_dim=num_latent, lengthscale=torch.ones(num_latent))
bmodel1= gp.models.VariationalSparseGP(X_init,x_train_torch.T,kernel1,Xu,likelihood1,whiten=True,jitter=1e-5)
bmodel2= gp.models.VariationalSparseGP(X_init,y_train_torch.T,kernel2,Xu,likelihood2,whiten=True,jitter=1e-5)
mrd=MRD(bmodel1,bmodel2)
mrd.to('cuda')
svi = SVI(mrd.model, mrd.guide, Adam({"lr":0.01, "betas":(0.95,0.999)}), TraceMeanField_ELBO())
for step in range(10000):
loss = svi.step()
if step % 100 == 0:
print("step {} loss = {:0.4g}".format(step, loss))
I want to estimate Y using Z (shown in the above image). In order to do this, I employed the following code to find X just using Z.
for param in mrd_model.parameters():
param.requires_grad_(False)
mrd_model.mode = "guide"
# fixes X
del mrd_model.X
mrd_model.X = mrd_model.X_loc.data
def model(y_new):
X_new = pyro.sample("X_new", dist.Normal(X_init, 1.0).to_event())
y_new_loc, y_new_var = mrd_model.predict_base_model1(X_new)
pyro.sample("y_new", dist.Normal(y_new_loc, y_new_var.sqrt()).to_event(), obs=y_new)
guide = autoguide.AutoDiagonalNormal(model)
svi = SVI(model, guide, Adam({"lr": 0.01, "betas": (0.95, 0.999)}), TraceMeanField_ELBO())
losses = []
y_new = x_train_torch.T
for i in range(50000):
loss = svi.step(y_new)
if i % 100 == 0:
print("step {} loss = {:0.4g}".format(i, loss))
losses.append(loss)
I was expecting when i consider the training data as the input. The above code could reproduce the same ( or at least close) hidden variables in the original model, but this didn’t happen.
If you visit this page, you can see that the similar code wasn’t able to reproduce the same hidden variables as expected.
Where do you think the problem came from?
Is it possible to use such model as regression tool as I want?
Thanks in advance