Hello,
I recently started using numpyro for my probabilistic programming on a project about data integration. However, I have a question about the label switching issue in the mixture model. I have read the existing posts about MM, but none of them can answer my question. I start a new post here. Sorry for the long wording.
The basic idea is as follows:
Suppose I have a pool of data from source subjects and data from one new subject that comes sequentially. Instead of using all of the data from the new subject all at once,
I would like to minimize the amount of data from the new subject by matching them with data from source subjects whenever additional sequence of data from new subject arrive.
During each sequence, there are 6 random vectors from two multivariate Gaussian distributions. Among 6 random vectors, five of them generate from a non-target MVN, and one of them generate from a target MVN.
It has a taste of a mixture model, but I do not have mixture weights for the data from the new subject because I assume it is from a baseline cluster. In addition, it seems to distinguish two MVNs across subject levels.
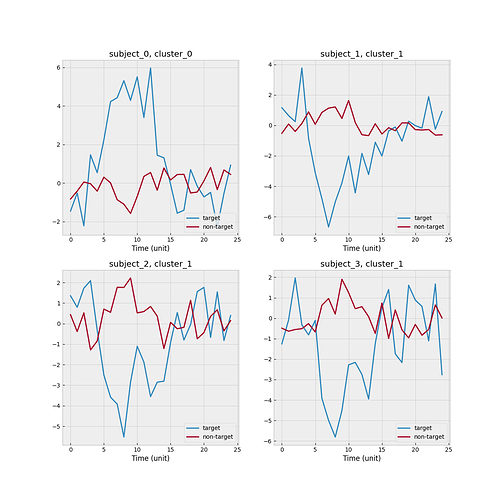
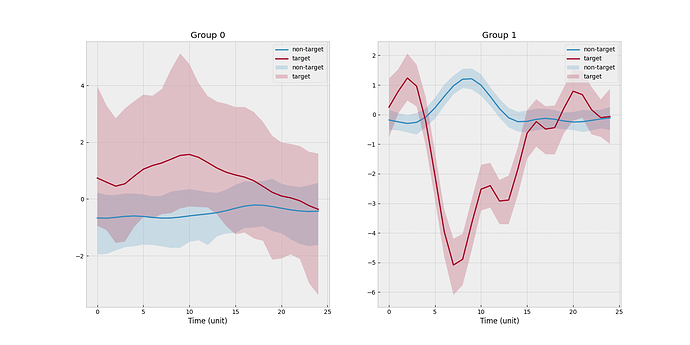
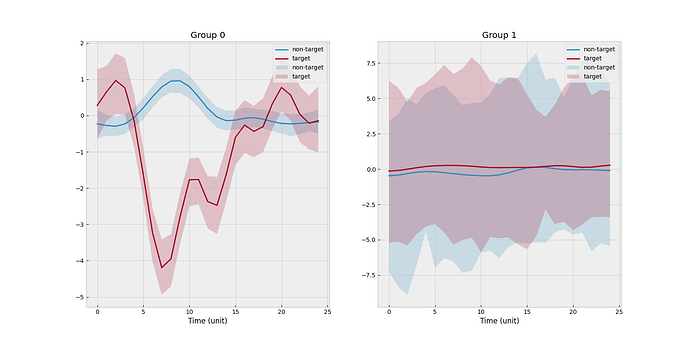
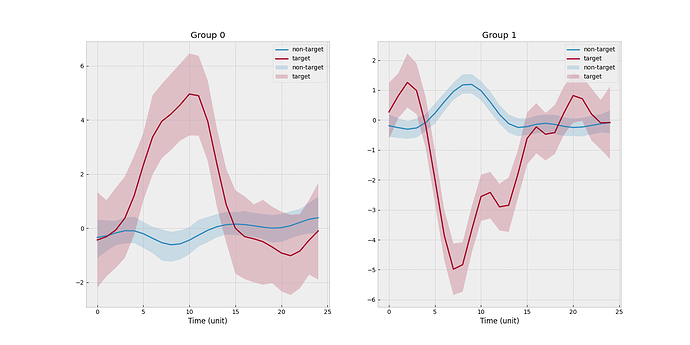
I create a toy example such that there are 2 clusters and 4 subjects. Subject 0 is the new subject, and cluster 0 is the one associated with the new subject. One scenario is that none of three source subjects match with the new subject. The empirical mean functions stratified by target/non-target and subject level is shown below.
The model code is also shown here:
def signal_integration_sim(
N, K, input_points=None, q_mat=None,
eigen_val_dict=None, eigen_fun_mat_dict=None,
input_data=None
):
signal_length = len(input_points)
var_ntar_dict = {}
var_tar_dict = {}
theta_ntar_dict = {}
theta_tar_dict = {}
beta_extend_dict = {}
s_sq_dict = {}
cov_mat_kron_dict = {}
for k in range(K):
group_k_name = 'group_{}'.format(k)
var_ntar_k_name = 'kernel_ntar_var_{}'.format(k)
var_ntar_k = numpyro.sample(var_ntar_k_name, dist.LogNormal(loc=0.0, scale=1.0))
var_ntar_dict[var_ntar_k_name] = var_ntar_k
theta_ntar_k_name = 'theta_ntar_{}'.format(k)
theta_ntar_k = numpyro.sample(
theta_ntar_k_name, dist.MultivariateNormal(
loc=jnp.zeros_like(eigen_val_dict[group_k_name]['non-target']),
covariance_matrix=jnp.diag(eigen_val_dict[group_k_name]['non-target'])
)
)
theta_ntar_dict[theta_ntar_k_name] = theta_ntar_k
beta_ntar_k = var_ntar_k * jnp.matmul(eigen_fun_mat_dict[group_k_name]['non-target'], theta_ntar_k)
var_tar_k_name = 'kernel_tar_var_{}'.format(k)
var_tar_k = numpyro.sample(var_tar_k_name, dist.LogNormal(loc=0.0, scale=1.0))
# var_tar_k = numpyro.sample(var_tar_k_name, dist.TruncatedNormal(loc=0.0, scale=5.0, low=0.0, high=10.0))
var_tar_dict[var_tar_k_name] = var_tar_k
theta_tar_k_name = 'theta_tar_{}'.format(k)
theta_tar_k = numpyro.sample(
theta_tar_k_name, dist.MultivariateNormal(
loc=jnp.zeros_like(eigen_val_dict[group_k_name]['target']),
covariance_matrix=jnp.diag(eigen_val_dict[group_k_name]['target'])
)
)
theta_tar_dict[theta_tar_k_name] = theta_tar_k
beta_tar_k = var_tar_k * jnp.matmul(eigen_fun_mat_dict[group_k_name]['target'], theta_tar_k)
beta_ls_k = jnp.stack([beta_tar_k, beta_ntar_k], axis=0)
beta_extend_k = jnp.reshape(
jnp.matmul(q_mat, beta_ls_k),
[rcp_unit_flash_num * signal_length]
)
beta_extend_k_name = 'beta_extend_{}'.format(k)
beta_extend_dict[beta_extend_k_name] = beta_extend_k
s_sq_k_name = 's_sq_{}'.format(k)
s_sq_k = numpyro.sample(s_sq_k_name, dist.HalfCauchy(scale=1.0))
s_sq_dict[s_sq_k_name] = s_sq_k
cov_mat_k = s_sq_k * jnp.eye(signal_length)
cov_mat_kron_k = jnp.kron(jnp.eye(rcp_unit_flash_num), cov_mat_k)
cov_mat_kron_k_name = 'cov_mat_kron_{}'.format(k)
cov_mat_kron_dict[cov_mat_kron_k_name] = cov_mat_kron_k
beta_extend_mix = jnp.stack([beta_extend_dict['beta_extend_{}'.format(k)] for k in range(K)], axis=0)
cov_mat_kron_mix = jnp.stack([cov_mat_kron_dict['cov_mat_kron_{}'.format(k)] for k in range(K)], axis=0)
# new subject only, I use the index from beta_extend_mix/cov_mat_kron_mix
beta_extend_0_name = 'beta_extend_0'
cov_mat_kron_0_name = 'cov_mat_kron_0'
data_0_name = 'subject_0'
new_0_dist = dist.MultivariateNormal(
loc=beta_extend_dict[beta_extend_0_name],
covariance_matrix=cov_mat_kron_dict[cov_mat_kron_0_name]
)
numpyro.sample(data_0_name, new_0_dist, obs=input_data[data_0_name])
# initialize the component distribution of the mixture model
component_dist = dist.MultivariateNormal(
loc=beta_extend_mix, covariance_matrix=cov_mat_kron_mix
)
for n in range(N-1):
prob_n_name = 'prob_{}'.format(n+1)
prob_n = numpyro.sample(prob_n_name, dist.Dirichlet(concentration=jnp.ones(K)))
mixing_n = dist.Categorical(probs=prob_n)
mixture_n_dist = dist.MixtureSameFamily(
mixing_n, component_dist
)
data_n_name = 'subject_{}'.format(n+1)
numpyro.sample(data_n_name, mixture_n_dist, obs=input_data[data_n_name])
Since I specify the distribution of data from subject 0 from parameters with index 0, group 0 and prob_x[0] are the parameters of interest.
In particular, prob_x[0] are expected to be small.
# new subject only, I use the index from beta_extend_mix/cov_mat_kron_mix
beta_extend_0_name = 'beta_extend_0'
cov_mat_kron_0_name = 'cov_mat_kron_0'
data_0_name = 'subject_0'
new_0_dist = dist.MultivariateNormal(
loc=beta_extend_dict[beta_extend_0_name],
covariance_matrix=cov_mat_kron_dict[cov_mat_kron_0_name]
)
numpyro.sample(data_0_name, new_0_dist, obs=input_data[data_0_name])
However, when I vary the sequence size of subject 0 (input_data['subject_0']), I got weird results.
When sequence size=1, the results make sense although the estimation of group 0 is bad (lack of sample size).
Partial summary table:
mean std median 5.0% 95.0% n_eff r_hat
kernel_ntar_var_0 0.73 0.35 0.68 0.27 1.29 217.73 1.00
kernel_ntar_var_1 0.87 0.24 0.83 0.49 1.18 101.82 1.03
kernel_tar_var_0 1.34 0.90 1.23 0.09 2.60 76.01 1.02
kernel_tar_var_1 2.69 0.55 2.61 1.87 3.62 51.98 1.04
prob_1[0] 0.08 0.06 0.06 0.00 0.16 306.61 1.00
prob_1[1] 0.92 0.06 0.94 0.84 1.00 306.61 1.00
prob_2[0] 0.08 0.07 0.06 0.00 0.18 291.58 1.00
prob_2[1] 0.92 0.07 0.94 0.82 1.00 291.58 1.00
prob_3[0] 0.08 0.07 0.06 0.00 0.18 194.66 1.00
prob_3[1] 0.92 0.07 0.94 0.82 1.00 194.66 1.00
s_sq_0 26.61 1.98 26.50 23.42 29.41 322.98 0.99
s_sq_1 25.91 0.38 25.90 25.26 26.48 301.55 1.00
However, for sequence size=3-6, the results seem to reverse, and the estimation for group 1 simply draws from the prior distributions. For example, given sequence size=3,
mean std median 5.0% 95.0% n_eff r_hat
kernel_ntar_var_0 0.76 0.24 0.72 0.39 1.01 131.75 1.00
kernel_ntar_var_1 1.78 2.42 0.94 0.05 4.21 229.30 1.01
kernel_tar_var_0 2.35 0.57 2.27 1.44 3.10 83.79 1.00
kernel_tar_var_1 1.71 2.27 0.96 0.07 3.99 200.75 1.00
prob_1[0] 0.92 0.07 0.94 0.81 1.00 514.90 1.00
prob_1[1] 0.08 0.07 0.06 0.00 0.19 514.90 1.00
prob_2[0] 0.91 0.09 0.94 0.79 1.00 278.67 1.00
prob_2[1] 0.09 0.09 0.06 0.00 0.21 278.67 1.00
prob_3[0] 0.92 0.08 0.95 0.79 1.00 208.25 1.00
prob_3[1] 0.08 0.08 0.05 0.00 0.21 208.25 1.00
When sequence size=7 and above, the results are normal.
mean std median 5.0% 95.0% n_eff r_hat
kernel_ntar_var_0 0.64 0.32 0.59 0.16 1.03 77.83 1.00
kernel_ntar_var_1 0.84 0.21 0.81 0.51 1.20 65.90 1.03
kernel_tar_var_0 2.62 0.70 2.54 1.70 3.76 52.75 1.00
kernel_tar_var_1 2.68 0.65 2.63 1.64 3.62 41.87 1.01
prob_1[0] 0.08 0.07 0.07 0.00 0.17 210.70 1.00
prob_1[1] 0.92 0.07 0.93 0.83 1.00 210.70 1.00
prob_2[0] 0.08 0.07 0.06 0.00 0.17 164.87 1.00
prob_2[1] 0.92 0.07 0.94 0.83 1.00 164.87 1.00
prob_3[0] 0.09 0.09 0.06 0.00 0.23 160.04 1.00
prob_3[1] 0.91 0.09 0.94 0.77 1.00 160.04 1.00
Since I used the Gaussian process to fit the curve, I have tried tuning the kernel hyper-parameters, increasing MCMC iteration size, changing the rng_key = random.PRNGKey(), unfortunately, none of them changed the inference results and the summary tables. I also checked the simulated data, I could spot the difference with my eyes.
Originally, I thought it was due to the label switching, but in my case, I already specified the label w.r.t the new subject and it was a two-cluster problem, so it should not cause any confusion.
Is it due to the label switching issue or the NUTS implementation?
Any feedback is appreciated.
Thanks,
Tianwen