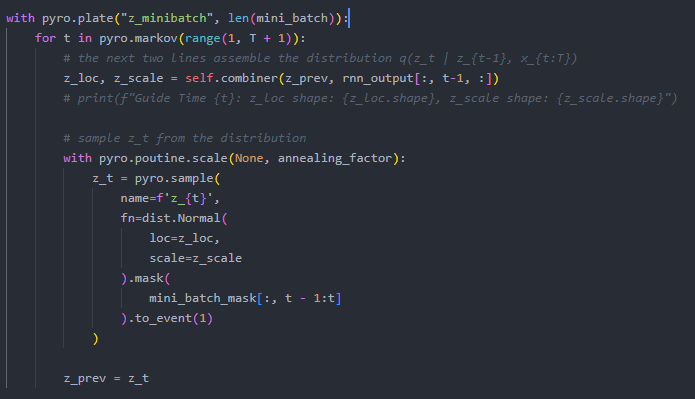
The first image shows the plate from my guide, and the second image is my model. The model’s goal is to observe the 1/0 label. In my guide, I’m using an RNN combined with latent states, similar to the DMM tutorial.
During the first iteration of training with SVI, the shapes of both parts across the timesteps are (batch_size, hidden_dim), which is correct for representing the latent vectors and distributions at each time step. However, after the first iteration, the sequence length is added to the first dimension (seq_len, batch_size, hidden_dim), and the dimensions alternate between being unsqueezed and squeezed: (seq_len, batch_size, hidden_dim) → (seq_len, 1, batch_size, hidden_dim) → (seq_len, batch_size, hidden_dim) → …
I’ve identified that this issue specifically occurs when checking the shape of the sampled latents from the second minibatch onward.
Could someone explain if my code is structured correctly or how I should format the plate/events to ensure no unwanted dimensions are added?
Thank you for any help!