I’m using an HMM to perform gesture recognition trained on several keypoints of the body. The model is something like
mu_i ~ N(0, 3)
sig_i ~ LogNormal(-3, 1)
transition_i ~ Dirichlet(15)
x_0 = 0
x_i ~ Cat(transition_{i-1})
y_i ~ N(mu_{x_i}, sig_{x_i})
However, when I train the HMM, the HMM ends throwing out all but one state. I’ve managed to repro the issue training just the emission center with a model as follows:
@config_enumerate
def model(data):
data = data.float()
num_cls = data.shape[0]
num_copies = data.shape[1]
num_dimensions = data.shape[3]
num_time = data.shape[2]
num_states = transition_matrix.shape[0]
with pyro.plate("num_cls", num_cls) as cls_idx:
with pyro.plate("hmm_gen", num_states - 1):
with pyro.plate("hmm_dim", num_dimensions):
emission_mus = pyro.sample('emission_mus',
distributions.Uniform(torch.tensor(-3.0), torch.tensor(3.0)))
with pyro.plate("copies", num_copies) as copies:
state = torch.zeros(num_copies, num_cls).long()
dimension_plate = pyro.plate("hmm_dim_gen", num_dimensions, dim=-3)
for t in pyro.markov(range(num_time)):
with dimension_plate as d:
emission_mu = Vindex(emission_mus)[d.unsqueeze(-1).unsqueeze(-1), state, cls_idx]
emission_sigma = Vindex(emission_sigmas)[d.unsqueeze(-1).unsqueeze(-1), state, cls_idx]
emission = pyro.sample("y_{}".format(t), distributions.Normal(emission_mu, emission_sigma),
obs=data[:, :, t].T)
state = pyro.sample("x_{}".format(t + 1),
distributions.Categorical(Vindex(transition_matrix)[state, cls_idx]))
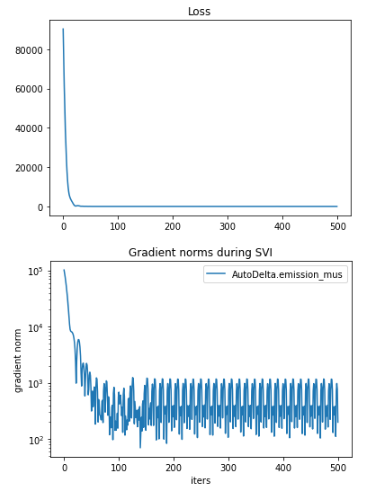
The full repro code can be seen at Full Code For Issue - Pastebin.com which also includes some sample data and the full training code. Anyway, when trained with this, it appears to train fine:
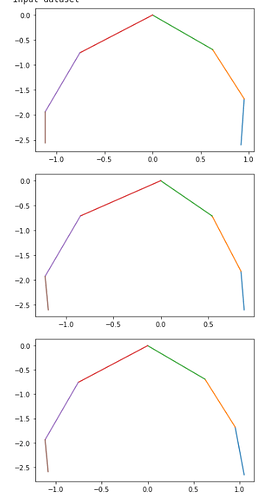
but, when the input dataset is visualized
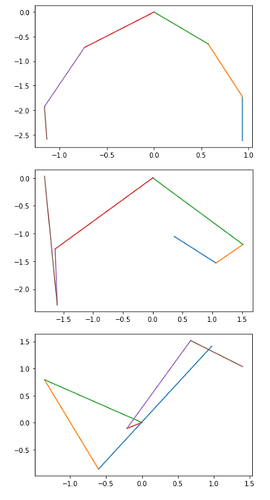
vs the learned center of the state emissions
You can observe that only the first state is used. This causes issues when using the model generatively as the transition matrix still transitions to the clusters which are not learned. I’m curious if this is the expected behavior of the SVI under the EnumElbo loss, and if so, if there’s any workaround to get all states to properly train? Thanks!