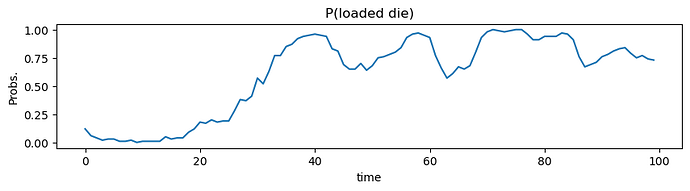
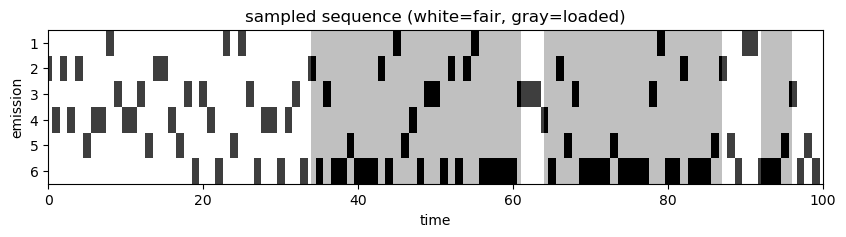
I am attempting to recreate the Dishonest Casino example from Kevin P. Murphy’s book “Advanced Probabilistic Machine Learning”. The authors offer an example notebook and solution in their new library Dynamax. However, I believe this can be recreated (and could be a nice example notebook) using Pyro’s discreteHMM distribution and use of the infer_discrete function. I am having trouble inferring the latent state for each observation (emission). The parameters for the HMM are:
initial_probs = torch.tensor([0.5, 0.5])
transition_matrix = torch.tensor([[0.95, 0.05],
[0.10, 0.90]])
emission_probs = torch.tensor([[1/6, 1/6, 1/6, 1/6, 1/6, 1/6], # fair die
[1/10, 1/10, 1/10, 1/10, 1/10, 5/10]]) # loaded die
Using the parameterised discreteHMM distribution, we can generate samples:
hmm = dist.DiscreteHMM(
initial_logits=torch.logit(initial_probs),
transition_logits=torch.logit(transition_matrix),
observation_dist=dist.Categorical(emission_probs),
duration=50
)
hmm.sample()
tensor([3, 2, 5, 5, 3, 5, 5, 5, 5, 5, 5, 3, 5, 0, 5, 5, 1, 5, 5, 1, 0, 1, 3, 1,
5, 5, 1, 1, 5, 1, 5, 0, 5, 5, 5, 5, 5, 3, 5, 5, 2, 1, 3, 5, 5, 5, 5, 0,
5, 2])
Each emission was generated according to some latent state (fair or loaded dice). Initially, I figured I could use the .filter() of the discreteHMM distribution to obtain the posterior of the latent state given a sequence of observations.
emissions = hmm.sample()
post_states = hmm.filter(emissions)
Here, the emissions are the sequence of observations generated by the HMM model. The post_states is a Categorical distribution. However, the .filter() method eliminates the time dimension, and therefore, post_states is the posterior over the final state given a sequence of observations (it does not return the most likely state that gave rise to each observation in the sequence). I would like to have the most probable state for each observation in the sequence using an inference technique like forward filtering or Viterbi (MAP).
This led me to discover the infer_discrete() function with the parameter temperature. When temperature is set to 1 this corresponds to forward filtering, and 0 corresponds to Viterbi-like MAP inference. Additionally, I read through the Inference with Discrete Latent Variables tutorial. Attempting to use infer_discrete standalone, I should be able to obtain MAP estimates for the latent state given a sequence of observations.
def model():
emissions = pyro.sample("emissions", dist.DiscreteHMM(
initial_logits=torch.logit(initial_probs),
transition_logits=torch.logit(transition_matrix),
observation_dist=dist.Categorical(emission_probs),
duration=50
), infer={"enumerate": "sequential"})
return emissions
serve = infer_discrete(model, first_available_dim=-1, temperature=0)
serve()
tensor([5, 5, 5, 3, 1, 2, 4, 1, 2, 0, 5, 5, 4, 5, 5, 5, 5, 2, 5, 5, 0, 5, 5, 5,
1, 1, 5, 5, 1, 1, 5, 1, 5, 5, 2, 5, 5, 5, 5, 4, 5, 5, 4, 5, 3, 5, 1, 5,
4, 3])
However, this only returns samples generated by the HMM model. Is it possible to use infer_discrete standalone to obtain MAP estimates of the latent states? Or will I need to define a model with priors and perform training in order to achieve this objective? I would like to have the posterior probability of each latent state that gave rise to the observation. In the tutorial linked above, there is a time series example, however, they do not use the discreteHMM distribution.
Thanks for your time and help. Cheers!