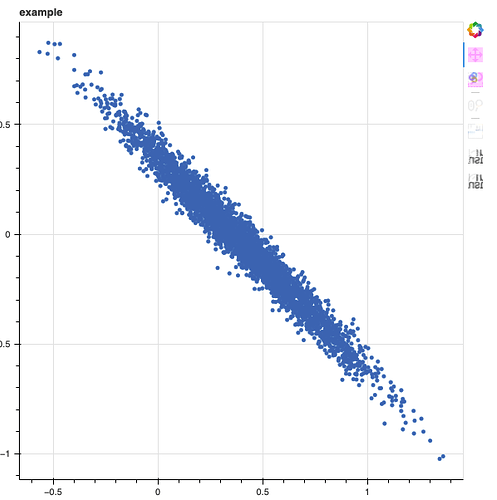
Suppose I have a 2-dimensional vector of linear model coefficients with independent Normal(0, 1) priors
theta = pyro.sample('theta', dist.Normal(torch.zeros(3), torch.ones(3)).to_event(1))
sigma = pyro.sample('sigma', dist.LogNormal(0, 1))
mu = torch.matmul(X, theta)
pyro.sample('obs', dist.Normal(mu, sigma), obs=y)
Is it safe to instead do
with pyro.plate("theta_plate", 3):
theta = pyro.sample("theta", Normal(0, 1)) # .expand([10]) is automatic
sigma = pyro.sample('sigma', dist.LogNormal(0, 1))
mu = torch.matmul(X, theta)
pyro.sample('obs', dist.Normal(mu, sigma), obs=y)
While the priors for each theta value are independent, the posterior for the theta values can still have correlation introduced by the likelihood. For example, if the columns of X are highly multicollinear.
I guess my fundamental question is what is pyro doing under the hood with plates? I’m having trouble understanding the precise meaning of the different independences. I’m coming from Stan, so I understand probabilistic modeling as specifying the joint log probability of any particular n-tuple of parameters, then running some kind of exploration algorithm over potential parameter tuples. What does it actually do to the log probability of a set of parameters when you run .to_event(1) vs looping over a plate?