When I calculated the WAIC with discrete latent variables, I found that the WAIC obtained by the Enumerated NUTS algorithm is much larger than that obtained by the Mixed HMC algorithm. Here is a toy model, and the code for data generation and parameter estimation is as follows:
import numpy as np
import jax
from jax import nn, random, vmap
import jax.numpy as jnp
import pandas as pd
import numpyro
from numpyro import handlers
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS, Predictive, DiscreteHMCGibbs, MixedHMC, HMC
import arviz as az
cat_p = np.random.uniform(0, 1)
cat = np.random.binomial(1, cat_p)
obs_p = np.array([0.95, 0.05])[cat]
Y = np.random.binomial(1,obs_p, 1000)
def tree_model():
cat_p = numpyro.sample("cat_p", dist.Beta(1,1))
cat = numpyro.sample("cat", dist.Bernoulli(cat_p), infer={"enumerate": "parallel"})
obs_p = jnp.array([0.95, 0.05])[cat]
with numpyro.plate("obs_plate", 1000, dim=-1):
obs = numpyro.sample("obs", dist.Bernoulli(obs_p), obs=Y)
def mixedHCM_tree():
cat_p = numpyro.sample("cat_p", dist.Beta(1,1))
cat = numpyro.sample("cat", dist.Bernoulli(cat_p))
obs_p = jnp.array([0.95, 0.05])[cat]
with numpyro.plate("obs_plate", 1000, dim=-1):
obs = numpyro.sample("obs", dist.Bernoulli(obs_p), obs=Y)
idatas = {}
kernel = NUTS(tree_model,)
mcmc = MCMC(kernel, num_warmup=20000, num_samples=10000, num_chains=4,)
mcmc.run(random.PRNGKey(1))
with numpyro.handlers.seed(rng_seed=1):
idatas["tree"] = az.from_numpyro(
mcmc,
posterior_predictive=pred
)
kernel = MixedHMC(HMC(mixedHCM_tree,))
mcmc = MCMC(kernel, num_warmup=20000, num_samples=10000, num_chains=4,)
mcmc.run(random.PRNGKey(1))
with numpyro.handlers.seed(rng_seed=1):
idatas["mixed_HMC_tree"] = az.from_numpyro(
mcmc,
posterior_predictive=pred
)
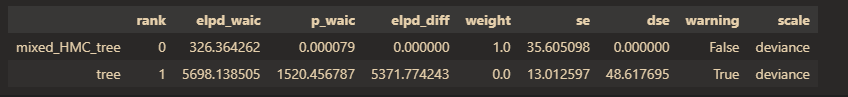
In theory, when estimating the same model with different estimation algorithms, the WAIC values should be similar. However, in the current results, the WAIC values differ greatly.
az.compare(idatas, scale="deviance", ic="waic")
Therefore, I would like to inquire why there is a significant difference in WAIC when using the Enumerated NUTS algorithm compared to using Mixed HMC algorithm. Is it a problem with the calculation of WAIC? If so, how can I correctly calculate fitting indicators such as -2LL and WAIC when using the Enumerated NUTS algorithm?
Thank you in advance. This means a lot to me.