Hi everyone,
i was reading the tutorial on BNNs implementation in pyro (Tutorial 1: Bayesian Neural Networks with Pyro — UvA DL Notebooks v1.2 documentation). In the first example, it is described how to use NUTS MCMC algorithm to sample from the posterior. Everything works ok but when I change the function and create bimodal function consisting of two sinusoids. Then my posterior predictive samples are not good.
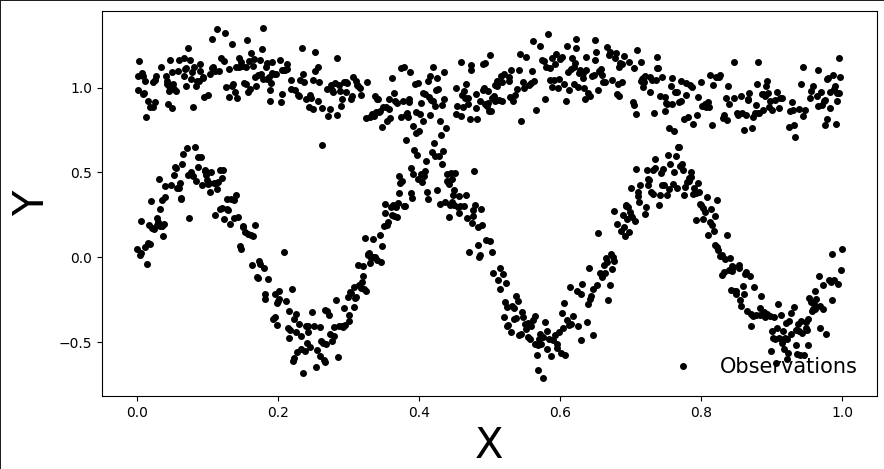
Here are the generated points of my bimodal distribution:
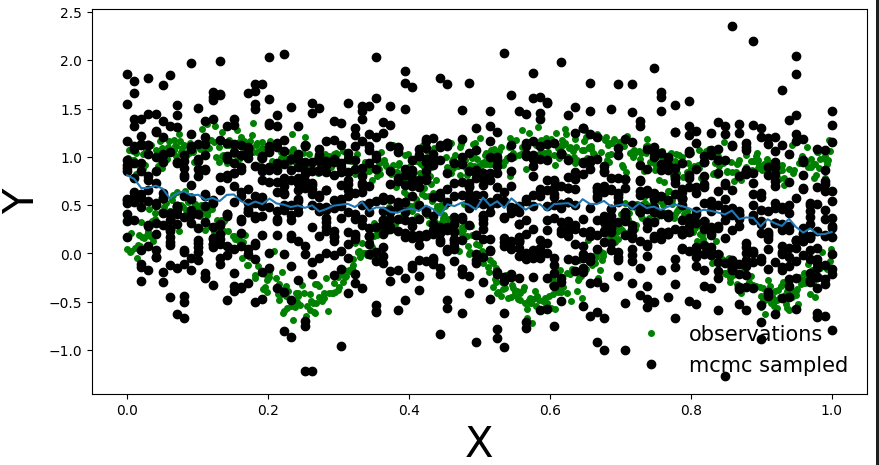
And here are the points from posterior predictive distribution. It looks that for each x the Y will be Gaussian.
Model is the following:
import pyro
import pyro.distributions as dist
from pyro.nn import PyroModule, PyroSample
import torch.nn as nn
class MyFirstBNN(PyroModule):
def __init__(self, in_dim=1, out_dim=1, hid_dim=5, prior_scale=10.):
super().__init__()
self.activation = nn.Tanh() # or nn.ReLU()
self.layer1 = PyroModule[nn.Linear](in_dim, hid_dim) # Input to hidden layer
self.layer2 = PyroModule[nn.Linear](hid_dim, out_dim) # Hidden to output layer
# Set layer parameters as random variables
self.layer1.weight = PyroSample(dist.Normal(0., prior_scale).expand([hid_dim, in_dim]).to_event(2))
self.layer1.bias = PyroSample(dist.Normal(0., prior_scale).expand([hid_dim]).to_event(1))
self.layer2.weight = PyroSample(dist.Normal(0., prior_scale).expand([out_dim, hid_dim]).to_event(2))
self.layer2.bias = PyroSample(dist.Normal(0., prior_scale).expand([out_dim]).to_event(1))
def forward(self, x, y=None):
x = x.reshape(-1, 1)
x = self.activation(self.layer1(x))
mu = self.layer2(x).squeeze()
sigma = pyro.sample("sigma", dist.Gamma(.5, 1)) # Infer the response noise
# Sampling model
with pyro.plate("data", x.shape[0]):
obs = pyro.sample("obs", dist.Normal(mu, sigma * sigma), obs=y)
return mu
I use NUTS to sample from posterior:
model = MyFirstBNN()
nuts_kernel = NUTS(model)
mcmc = MCMC(nuts_kernel, warmup_steps=100,num_samples=300)
# Convert data to PyTorch tensors
x_train = torch.from_numpy(x_obs).float().unsqueeze(1)
y_train = torch.from_numpy(y_obs).float()
# Run MCMC
mcmc.run(x_train, y_train)
And I generate posterior predictive using the following code:
predictive = Predictive(model=model, posterior_samples=mcmc.get_samples(), return_sites=("obs", "_RETURN"))
x_test = torch.linspace(0,1, 100)
preds = predictive(x_test.unsqueeze(1))
Am I doing something wrong (too small number of samples, maybe there is some other NUTS settings) or it is just impossible to use BNN with Gaussian posterior predictive for such cases? I was reading paper Hands-on Bayesian Neural Networks – a Tutorial for Deep Learning Users
([2007.06823] Hands-on Bayesian Neural Networks -- a Tutorial for Deep Learning Users) which actually claims that it should be possible with MCMC BNN.
PS. If I add only one mode (one sinusoid instead of two), then the posterior predictive fit is good.
Thank you in advance!
Best