The diagnostic classification tree model is a class of educational measurement models used to model student responses with a response matrix of N rows and I columns, where N represents the number of students and I represents the number of questions (items).
As shown in the figure, the first node indicates whether student n exhibits guessing behavior on the question i. When guessing behavior is exhibited, the probability of a correct answer for subject n on question i is 0.25. When the student does not exhibit guessing behavior, the probability of a correct answer to question i is determined according to the category in which student n is placed, e.g., when the student is in category I, the probability of a correct answer for the student is 0.9.
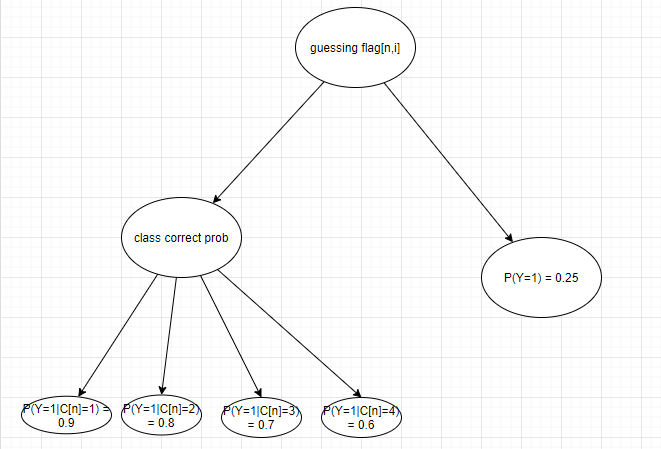
Based on this model in the figure, I converted it into a code for numpyro, however, this code encountered problems and reported errors when enumerating discrete variables.
import numpy as np
from jax import nn, random, vmap
import jax.numpy as jnp
import numpyro
from numpyro import handlers
import numpyro.distributions as dist
from numpyro.infer import MCMC, NUTS, Predictive, DiscreteHMCGibbs, MixedHMC, HMC
from numpyro.infer.reparam import LocScaleReparam
from numpyro.ops.indexing import Vindex
import itertools
N,I = 20, 5
Y = np.random.binomial(1,0.5, (20,5))
# with numpyro.handlers.seed(rng_seed=0):
def m():
student_plate = numpyro.plate("student", N, dim=-2)
item_plate = numpyro.plate("item", I, dim=-1)
# Assumpation there are 4 class of student, and the correct of each class on each item is known
classs_correct_prob = jnp.stack([jnp.array([0.9,0.8,0.6,0.5])
for i in range(I)]).T # <---- shape = (class_count, item_count)
class_prevalence = jnp.repeat(0.25,4)
with student_plate:
# sample class of student
student_c = numpyro.sample("cat", dist.Categorical(class_prevalence),
infer={"enumerate": "parallel"})
# prior of student is flaged as guessing
gussing_flag_prior = numpyro.sample("gussing_flag_prior", dist.Beta(1,5))
with item_plate:
# get the flag of rg
gussing_flag_idx = numpyro.sample("gussing_flag_idx",
dist.Bernoulli(gussing_flag_prior),
infer={"enumerate": "parallel"})
gussing_flag = Vindex(jnp.asarray([0,1]))[gussing_flag_idx]
# correct prob for student on item if student not guessing
c_p_not_g = Vindex(classs_correct_prob)[student_c]
# correct prob consider gussing flag
correct_prob = 0.25*gussing_flag + c_p_not_g.squeeze()*(1-gussing_flag)
numpyro.sample("Y",dist.Bernoulli(probs=correct_prob.squeeze()), obs=Y)
ERROR
Output exceeds the size limit. Open the full output data in a text editor
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
[... skipping hidden 1 frame]
File ~/anaconda3/envs/pymc3/lib/python3.10/site-packages/jax/_src/util.py:219, in cache.<locals>.wrap.<locals>.wrapper(*args, **kwargs)
218 else:
--> 219 return cached(config._trace_context(), *args, **kwargs)
File ~/anaconda3/envs/pymc3/lib/python3.10/site-packages/jax/_src/util.py:212, in cache.<locals>.wrap.<locals>.cached(_, *args, **kwargs)
210 @functools.lru_cache(max_size)
211 def cached(_, *args, **kwargs):
--> 212 return f(*args, **kwargs)
File ~/anaconda3/envs/pymc3/lib/python3.10/site-packages/jax/_src/lax/lax.py:126, in _broadcast_shapes_cached(*shapes)
124 @cache()
125 def _broadcast_shapes_cached(*shapes: Tuple[int, ...]) -> Tuple[int, ...]:
--> 126 return _broadcast_shapes_uncached(*shapes)
File ~/anaconda3/envs/pymc3/lib/python3.10/site-packages/jax/_src/lax/lax.py:142, in _broadcast_shapes_uncached(*shapes)
141 if result_shape is None:
--> 142 raise ValueError("Incompatible shapes for broadcasting: {}"
143 .format(tuple(shape_list)))
144 return result_shape
ValueError: Incompatible shapes for broadcasting: ((1, 20, 5), (2, 4, 5))
...
--> 142 raise ValueError("Incompatible shapes for broadcasting: {}"
143 .format(tuple(shape_list)))
144 return result_shape
ValueError: Incompatible shapes for broadcasting: ((1, 20, 5), (2, 4, 5))
I am confused by this error because I can’t find the array of this shape.
with numpyro.handlers.seed(rng_seed=0):
trace = numpyro.handlers.trace(m).get_trace()
print(numpyro.util.format_shapes(trace))
How can I use numpyro to marginalize a discrete variable in the tree model and implement the estimation of the parameters?
Any advice or reference will be appreciated~