Hi Numpyro fans,
I have a 21 parameters sampling to manage and I am using Numpyro NUTS with a model consisting of Uniform priors for same variables and Normal priors for the others, and finally the likelihood is a MultivariateNormal(signal, cov) distribution where cov is a constant covariance matrix 2250 x 2250. Now,
I use a simple running
nuts_kernel = numpyro.infer.NUTS(cond_model,
init_strategy=numpyro.infer.init_to_sample(),
max_tree_depth=5) # max_tree_depth=10
mcmc = numpyro.infer.MCMC(nuts_kernel,
num_warmup=1_000,
num_samples=8_000,
num_chains=1,
jit_model_args=True,
progress_bar=False)
The job summary indicates that r_hat is 1.00-1.01 while the n_eff for some variables are O(150) but some most are about 50 which is rather low effeiciency.
I have identified that 5 variables are highly correlated.
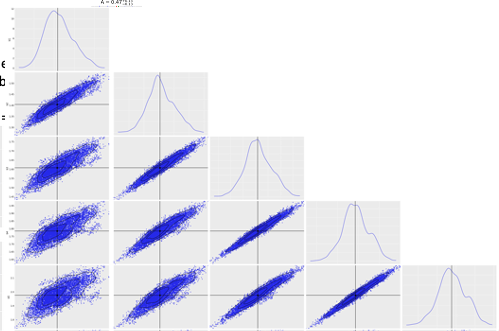
Do you have some advises to make the sampling more efficient? what about the mass matrix shaping for instance ?